A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Quantification of Ethanol Levels in Zebrafish Embryos Using Head Space Gas Chromatography
In This Article
Summary
This work describes a protocol to quantify ethanol levels in a zebrafish embryo using head space gas chromatography from proper exposure methods to embryo processing and ethanol analysis.
Abstract
Fetal Alcohol Spectrum Disorders (FASD) describe a highly variable continuum of ethanol-induced developmental defects, including facial dysmorphologies and neurological impairments. With a complex pathology, FASD affects approximately 1 in 100 children born in the United States each year. Due to the highly variable nature of FASD, animal models have proven critical in our current mechanistic understanding of ethanol-induced development defects. An increasing number of laboratories has focused on using zebrafish to examine ethanol-induced developmental defects. Zebrafish produce large numbers of externally fertilized, genetically tractable, translucent embryos. This allows researchers to precisely control timing and dosage of ethanol exposure in multiple genetic contexts and quantify the impact of embryonic ethanol exposure through live imaging techniques. This, combined with the high degree of conservation of both genetics and development with humans, has proven zebrafish to be a powerful model in which to study the mechanistic basis of ethanol teratogenicity. However, ethanol exposure regimens have varied between different zebrafish studies, which has confounded the interpretation of zebrafish data across these studies. Here is a protocol to quantify ethanol concentrations in zebrafish embryos using head space gas chromatography.
Introduction
Fetal Alcohol Spectrum Disorders (FASD) describes a wide array of neurological impairments and craniofacial dysmorphologies associated with embryonic ethanol exposure1. Multiple factors, including timing and dosage of ethanol exposure and genetic background, contribute to the variation of FASD2,3. In humans, the complex relationship of these variables makes studying and understanding the etiology of FASD challenging. Animal models have proven crucial in developing our understanding of the mechanistic basis of ethanol teratogenicity. A wide variety of animal model systems has been used to study multiple aspects of FASD and results have been remarkably consistent with what is found in exposure in humans4. Rodent model systems are used to examine many aspects of FASD, with mice being the most common5,6,7. The majority of this work has focused on developmental defects to early ethanol exposure8, though later exposure to ethanol has been shown to cause developmental anomalies as well9. Moreover, the genetic capabilities of mice have greatly aided in our ability to probe the genetic underpinnings of FASD10,11. These studies in mice strongly suggest that there are gene-ethanol interactions with the sonic hedgehog pathway, retinoic acid signaling, Superoxide dismutase, Nitric oxide synthase I, Aldh2 and Fancd28,10,11,12,13,14,15,16,17,18,19,20,21. These studies show that animal models are critical to advancing our understanding of FASD and its underlying mechanisms.
The zebrafish has emerged as a powerful model system to examine many aspects of ethanol teratogenesis22,23. Due to their external fertilization, high fecundity, genetic tractability, and live imaging capabilities, zebrafish are ideally suited to study factors such as timing, dosage, and genetics of ethanol teratogenesis. Ethanol can be administered to precisely staged embryos and the embryos can then be imaged to examine the direct impact of ethanol during developmental processes. This work can be related directly to humans, because the genetic programs of development are highly conserved between zebrafish and humans and can therefore help guide FASD human studies24. While zebrafish have been used to examine ethanol teratogenesis, a lack of consensus in reporting embryonic ethanol concentrations makes comparison to humans difficult25. In mammalian systems, blood-alcohol levels correlate directly to tissue ethanol levels26. Many of the zebrafish studies treat embryos before complete formation of their circulatory system. With no maternal sample to examine, a process to assess ethanol concentrations is required to quantify ethanol levels within the embryo. Here we describe a process to quantify ethanol concentrations in a developing zebrafish embryo using head space gas chromatography.
Access restricted. Please log in or start a trial to view this content.
Protocol
All zebrafish embryos used in this procedure were raised and bred following established IACUC protocols27. These protocols were approved by the University of Texas at Austin and the University of Louisville.
NOTE: The zebrafish line Tg(fli1:EGFP)y1 was used in this study28. All water used in this procedure is sterile reverse osmosis water. All statistical analyses were performed using Graphpad Prism v8.2.1.
1. Making embryo media
- To make a 20x stock of embryo media, dissolve 17.5 g of NaCl, 0.75 g of KCl, 2.9 g of CaCl2, 0.41 g of K2HPO4, 0.142 g of NA2HPO4, and 4.9 g of MgSO4·7H2O in 1 L of water. Ignore the white precipitate that forms; this will not impact the media. Filter sterilize the stock solution and store at 4 °C.
- To create the working embryo media solution, dissolve 1.2 g of NaHCO3 in 1 L of 20x embryo media stock and add 19 L of water. Maintain the working embryo media solution at 28 °C.
2. Measuring the embryonic volume using water displacement
NOTE: In this protocol, 24 h postfertilization (hpf) embryos (Figure 1) are used. The embryos used in the volume measurements are not used in the ethanol analysis.
- Place 10 embryos and extraembryonic fluid in 1.5 mL microcentrifuge tube marked at a volume of 250 μL (Figure 2A). Add water to the 250 μL fill line (see sample with dyed water in Figure 2B).
- Repeat step 2.1 to set up receptacles for embryos that have had their chorions removed.
- To remove the chorion, place the embryos in their chorions in a 100 mm Petri dish with 2 mg/mL of protease cocktail in embryo media at room temperature (RT) for 10 min. Every few minutes, gently swirl the embryos to break the chorion.
- Once all the embryos are free of their chorion, remove the dechorionated embryos from protease cocktail/embryo media and place in a new 100 mm Petri dish with fresh embryo media to wash the embryos. Repeat this wash step 1 more time (for a total of 2 washes). Transfer the embryos to a fresh 100 mm Petri dish.
- Using the smallest tip possible and without damaging the embryos, carefully remove all liquid from around the embryos using a p200 micropipettor (Figure 2C) and weigh the water with a scale with <0.1 mg precision. To determine the volume of the sample of 10 embryos, subtract the weight/volume of the water (1 mL of water = 1 g of water.) removed from 250 μL. To determine the volume of a single embryo, divide the difference between 250 μL and the weight of the water removed by 10.
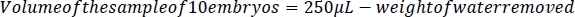
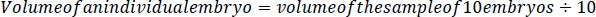
3. Treating embryos with ethanol
- Gather embryos from mating tanks and place in a standard 100 mm Petri dish. Count out no more than 100 embryos per single Petri dish and incubate at 28.5 °C.
NOTE: If the chorion is to be removed (step 2.2), it needs to be removed before adding ethanol. - At 6 hpf, add up to 100 embryos to a new standard 100 mm Petri dish either with embryo media or embryo media + 1% ethanol (v/v). Cover, but DO NOT seal the Petri dish. Place the embryos in a low temp incubator set at 28.5 °C for 18 h, or until the embryos reach the developmental time point of 24 hpf.
4. Preparing workflow before processing the embryos for head space gas chromatography
- Make a solution of 5 M NaCl in water (450 μL will be needed per sample tube) to denature all proteins and prevent ethanol metabolism. Make the protease cocktail (Table of Materials) at a concentration of 2 mg/mL in embryo media.
- Label a 1.5 mL microcentrifuge tube and a 2 mL gas chromatograph vial for each sample. Label additional 2 mL gas chromatograph vials for the ethanol standards described below as well as air, water, and the 5 M NaCl/protease cocktail blanks.
- Set up three p200 micropipettors two set to 50 μL, and the third set to 200 μL; a p1000 micropipettor set to 450 μL and a p2 micropipettor set to 2 μL. For the glass pipettes used to transfer embryos from the Petri dishes to the 1.5 mL microcentrifuge tubes, quickly pass the pipette tip through a flame to smooth the edges to not damage the embryos when drawing them into the pipette.
5. Processing embryos for head space gas chromatography
NOTE: Both embryos in their chorions and those previously removed from their chorions are treated the same for consistency in the calculation of dilution factors.
- Using the two p200 micropipettors set to 50 μL, draw 50 μL of the protease cocktail solution into one and draw 50 μL of water into the second.
- Using the glass pipette and micropipettor, quickly place 10 embryos (from step 3.2) in a 1.5 mL microcentrifuge tube and close the cap (as described in Figure 2A). Repeat for all samples to be tested, controls, and ethanol treated embryos.
- Using the p200 micropipettor set to 200 μL, quickly open the cap and remove all residual embryo media (as described in Figure 2C). Quickly place the pipette containing 50 μL of water in the tube and rapidly but gently (to not damage the embryos) add, then remove the water. Quickly add 50 μL of the protease cocktail solution from the waiting pipettor and close the tube cap (Figure 2D).
NOTE: Perform this process one sample at a time. - Let the sample sit at RT for 10 min to allow the protease cocktail to degrade the chorion. Then, quickly add 450 μL of 5 M NaCl and close the tube cap (Figure 2E). Vortex the samples for 10 min. To speed up the process, set up the mixer to vortex multiple samples at the same time.
NOTE: Add a small amount (~100 μL) of a silica bead mixture (2 sizes, 0.5 mm and 1 mm bead) to any tube with embryos older than 24 hpf. In older embryos, the notochord will remain intact regardless of how long they are homogenized. - After homogenizing for 10 min, quickly remove 2 μL of homogenized embryo supernatant and add to a gas chromatograph vial. Quickly seal the vial with the polytetrafluoroethylene cap.
6. Preparing media and ethanol standards
- To prepare the media standards, dilute the media by a factor of 10 with a 5 M NaCl/protease cocktail solution. Add 2 μL of each sample to a gas chromatograph vial and seal with a polytetrafluoroethylene cap.
- To prepare the ethanol standards, create a serial dilution of 100% ethanol in 5 M NaCl/protease cocktail solution to the following concentrations: 0.3125, 0.625, 1.25, 2.5, 5, 10, 20, and 40 mM. Add 2 μL of each standard to a gas chromatograph vial and seal with a polytetrafluoroethylene cap.
7. Preparing the head space gas chromatograph
NOTE: This setup and protocol may need to be changed depending on the gas chromatograph used. Head space gas chromatography is used to quantify ethanol levels, not for separation.
- Set the heater for the autosampler to 58 °C and turn on. Allow the heater to reach 58 °C, and turn on the air and hydrogen gas lines feeding the gas chromatograph (for flame ionization used to quantify the ethanol).
NOTE: The psi should be set to properly operate the chromatograph according to the manufacturer's specifications. Make sure the helium line is on and set to the proper psi. - For the septum, make sure the number of samples injected is not >100. For the injection fiber, make sure the number of samples injected is not >500.
NOTE: If either is over the amount of injections, they will need to be changed before running the test samples. - Turn on the analysis software and make sure the workstation is set up properly. Store all data according to lab/department standards. Create a new sample list for the samples to be run.
- Initiate the startup method and wait for the flame ionization detector to stabilize before running the samples. Once stable, run the software startup method to clean the fiber.
8. Sample measurements using head space gas chromatography
- Once the startup method is complete, create 1 new line per sample in the sample list. From the methods menu in the analysis software, load 436 Current spme ethanol 2013 3min absorb 2_5min rg run.METH for each sample on the sample list.
- Fill out the sample list, starting with the air, water, 5 M NaCl/protease cocktail blanks. Then enter in the standards in order, from 0.3125 to 40 mM. Follow the standards with a second round of the air, water, and 5 M NaCl/protease cocktail blanks.
- Enter all homogenized embryo supernatant samples to be tested, from lowest to highest predicted ethanol concentration. End by entering a third and final round of the air, water, and 5 M NaCl/protease cocktail blanks.
- Add the gas chromatograph vials to the autosampler in the order in which the samples were entered. Allow samples to warm for 10–15 min. Start the sample runs in the software.
- After all the samples and the final blanks have run, activate the shutdown method in the software by adding a final sample in the sample list and running standby.METH. Back up all data acquired during the sample runs. Turn off the equipment in the following order: autosampler heater, hydrogen tank and, only after the chromatograph temperature reaches 30 °C, the air tank. Leave the helium tank on to preserve the wax column.
9. Sample ethanol peak integration and sample concentration analysis
NOTE: All values from 9.3 on were calculated in an excel file that all equations prefilled.
- Once the shutdown method is complete, click on Open chromatogram. Open the folder containing the results. Samples are automatically integrated in this program.
- In the results, make sure the correct peaks have been integrated (ethanol peaks between 2 and 2.5). Once all samples have been confirmed, print or export the results.
- Plot the peak height of the ethanol standards on a graph. Calculate the slope, Y intercept and R2 values for the ethanol standards (R2 should be >0.99).
NOTE: These values will be used to determine the ethanol concentration from the sample peak height. - For each sample, subtract the peak height of the sample from the Y intercept of the ethanol standards. Divide this value by the slope of the ethanol standards to obtain the ethanol value for each sample in the GC vials.
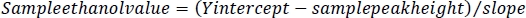
- To calculate the ethanol concentration in the embryos, first calculate the dilution factor for each sample. Take the volume measure calculated in step 2.3 of this protocol and divide it by the volume measure plus 500 μL. This represents the 5 M NaCl/protease cocktail solution added to each sample during embryo processing (steps 5.3 and 5.4).
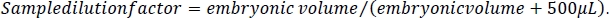
- Using this sample dilution factor, multiply by the sample ethanol value for each sample. The results will be in mM concentration. Calculate media reference samples by multiplying the media ethanol value by the dilution factor of 10.
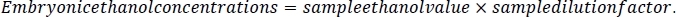
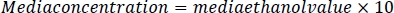
Access restricted. Please log in or start a trial to view this content.
Results
Blood ethanol levels cannot be determined in early embryonic zebrafish, because they lack a fully formed circulatory system. To determine the level of ethanol concentration in the zebrafish embryos, the ethanol levels are measured directly from homogenized embryonic tissue. To properly measure the embryonic ethanol concentrations, the embryonic volume has to be taken into account. The embryo (yolk attached) sits inside the chorion (eggshell) surrounded by extraembryonic fluid (Figure 1). Any...
Access restricted. Please log in or start a trial to view this content.
Discussion
As a developmental model system, zebrafish are ideally suited to study the impact of environmental factors on development. They produce large numbers of externally fertilized embryos, which allows for precise timing and dosage paradigms in ethanol studies. This, combined with the live imaging capabilities and the genetic and developmental conservation with humans, make zebrafish a powerful model system for teratology studies. Described is a protocol for measuring embryonic ethanol concentrations in developing zebrafish e...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The research presented in this article was supported by previous grants from National Institutes of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) R01DE020884 to J.K.E. and National Institutes of Health/National Institute on Alcohol Abuse and Alcoholism (NIH/NIAAA) F32AA021320 to C.B.L. and by the current grant from National Institutes of Health/National Institute on Alcohol Abuse (NIH/NIAAA) R00AA023560 to C.B.L. We thank Rueben Gonzales for providing and assisting with gas chromatograph analysis. We thank Tiahna Ontiveros and Dr. Gina Nobles writing assistance.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Air | Provided by contract to the university | ||
| Analytical Balance | VWR | 10204-962 | |
| AutoSampler, CP-8400 | Varian | Gas Chromatograph Autosampler | |
| Calcium Chloride | VWR | 97062-590 | |
| Ethanol | Decon Labs | 2701 | |
| Gas chromatograph vial with polytetrafluoroethylene/silicone septum and plastic cap 2 mL | Agilent | 8010-0198 | Can reuse the vials after cleaning, but not the caps/septa |
| Gas Chromatograph, CP-3800 | Varian | ||
| Helium | Provided by contract to the university | ||
| HP Innowax capillary column | Agilent | 19095N-123I | 30 m x 0.53 mm x 1.0 μm film thick |
| Hyrdogen | Provided by contract to the university | ||
| Magnesium Sulfate (Heptahydrate) | Fisher Scientific | M63-500 | |
| Microcentrifuge tube 1.5 mL | Fisher Scientific | 2682002 | |
| Micropipette tips 10 μL | Fisher Scientific | 13611106 | |
| Micropipette tips 1000 μL | Fisher Scientific | 13611127 | |
| Micropipette tips 200 μL | Fisher Scientific | 13611112 | |
| Petri dishes 100 mm | Fisher Scientific | FB012924 | |
| Pipetman L p1000L Micropipette | Gilson | FA10006M | |
| Pipetman L p200L Micropipette | Gilson | FA10005M | |
| Pipetman L p2L Micropipette | Gilson | FA10001M | |
| Polytetrafluoroethylene/silicone septum and plastic cap | Agilent | 5190-7021 | Replacement caps/septa for gas chromatograph vials |
| Potassium Chloride | Fisher Scientific | P217-500 | |
| Potassium Phosphate (Dibasic) | VWR | BDH9266-500G | |
| Pronase | VWR | 97062-916 | |
| Silica Beads .5 mm | Biospec Products | 11079105z | |
| Silica Beads 1.0 mm | Biospec Products | 11079110z | |
| Sodium Bicarbonate | VWR | BDH9280-500G | |
| Sodium Chloride | Fisher Scientific | S271-500 | |
| Sodium Phosphate (Dibasic) | Fisher Scientific | S374-500 | |
| Solid-phase microextraction fiber assembly Carboxen/Polydimethylsiloxane | Millipore Sigma | 57343-U | Replacement fibers |
| Star Chromatography Workstation | Varian | Chromatography software | |
| Thermogreen Low Bleed (LB-2) Septa | Millipore Sigma | 23154 | Replacement inlet septa |
References
- Elliott, E. J., Payne, J., Morris, A., Haan, E., Bower, C. Fetal alcohol syndrome: a prospective national surveillance study. Archive of Diseases in Childhood. 93 (9), 732-737 (2008).
- Cudd, T. A. Animal model systems for the study of alcohol teratology. Experimental Biology and Medicine. 230 (6), 389-393 (2005).
- Williams, J. F., Smith, V. C. Committee on Substance Abuse. Fetal Alcohol Spectrum Disorders. Pediatrics. 136 (5), 1395-1406 (2015).
- Patten, A. R., Fontaine, C. J., Christie, B. R. A comparison of the different animal models of fetal alcohol spectrum disorders and their use in studying complex behaviors. Frontiers in Pediatrics. 2, 93(2014).
- Petrelli, B., Weinberg, J., Hicks, G. G. Effects of prenatal alcohol exposure (PAE): insights into FASD using mouse models of PAE. Biochemistry and Cell Biology. 96 (2), 131-147 (2018).
- Mayfield, J., Arends, M. A., Harris, R. A., Blednov, Y. A. Genes and Alcohol Consumption: Studies with Mutant Mice. International Review Neurobiology. 126, 293-355 (2016).
- Marquardt, K., Brigman, J. L. The impact of prenatal alcohol exposure on social, cognitive and affective behavioral domains: Insights from rodent models. Alcohol. 51, 1-15 (2016).
- Sulik, K. K. Genesis of alcohol-induced craniofacial dysmorphism. Experimental Biology and Medicine. 230 (6), 366-375 (2005).
- Lipinski, R. J., et al. Ethanol-induced face-brain dysmorphology patterns are correlative and exposure-stage dependent. PLoS One. 7 (8), 43067(2012).
- Eberhart, J. K., Parnell, S. The genetics of fetal alcohol spectrum disorders. Alcoholism: Clinical and Experimental Research. 40 (6), 1154-1165 (2016).
- Becker, H. C., Diaz-Granados, J. L., Randall, C. L. Teratogenic actions of ethanol in the mouse: a minireview. Pharmacology, Biochemistry and Behavior. 55 (4), 501-513 (1996).
- Ahlgren, S. C., Thakur, V., Bronner-Fraser, M. Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proceedings of the National Academy of Sciences of the United States of America. 99 (16), 10476-10481 (2002).
- Loucks, E. J., Ahlgren, S. C. Deciphering the role of Shh signaling in axial defects produced by ethanol exposure. Birth Defects Research Part A: Clinical and Molecular Teratology. 85 (6), 556-567 (2009).
- Hong, M., Krauss, R. S. Cdon mutation and fetal ethanol exposure synergize to produce midline signaling defects and holoprosencephaly spectrum disorders in mice. PLoSGenetics. 8 (10), 1002999(2012).
- Aoto, K., Shikata, Y., Higashiyama, D., Shiota, K., Motoyama, J. Fetal ethanol exposure activates protein kinase A and impairs Shh expression in prechordal mesendoderm cells in the pathogenesis of holoprosencephaly. Birth Defects Research Part A: Clinical and Molecular Teratology. 82 (4), 224-231 (2008).
- Deltour, L., Ang, H. L., Duester, G. Ethanol inhibition of retinoic acid synthesis as a potential mechanism for fetal alcohol syndrome. The FASEB Journal. 10 (9), 1050-1057 (1996).
- Wentzel, P., Eriksson, U. J. Ethanol-induced fetal dysmorphogenesis in the mouse is diminished by high antioxidative capacity of the mother. Toxicological Sciences. 92 (2), 416-422 (2006).
- Karacay, B., Mahoney, J., Plume, J., Bonthius, D. J. Genetic absence of nNOS worsens fetal alcohol effects in mice. II: microencephaly and neuronal losses. Alcoholism: Clinical and Experimental Research. 39 (2), 221-231 (2015).
- Bonthius, D. J. Jr, Winters, Z., Karacay, B., Bousquet, S. L., Bonthius, D. J. Importance of genetics in fetal alcohol effects: null mutation of the nNOS gene worsens alcohol-induced cerebellar neuronal losses and behavioral deficits. Neurotoxicology. 46, 60-72 (2015).
- Bonthius, D. J., et al. Deficiency of neuronal nitric oxide synthase (nNOS) worsens alcohol-induced microencephaly and neuronal loss in developing mice. Brain Research. Developmental Brain Research. 138 (1), 45-59 (2002).
- Langevin, F., Crossan, G. P., Rosado, I. V., Arends, M. J., Patel, K. J. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 475 (7354), 53-58 (2011).
- Lovely, C. B., Fernandes, Y., Eberhart, J. K. Fishing for Fetal Alcohol Spectrum Disorders: Zebrafish as a Model for Ethanol Teratogenesis. Zebrafish. 13 (5), 391-398 (2016).
- Fernandes, Y., Buckley, D. M., Eberhart, J. K. Diving into the world of alcohol teratogenesis: a review of zebrafish models of fetal alcohol spectrum disorder. Biochemistry and Cell Biology. 96 (2), 88-97 (2018).
- McCarthy, N., et al. Pdgfra protects against ethanol-induced craniofacial defects in a zebrafish model of FASD. Development. 140 (15), 3254-3265 (2013).
- Lovely, C. B., Nobles, R. D., Eberhart, J. K. Developmental age strengthens barriers to ethanol accumulation in zebrafish. Alcohol. 48 (6), 595-602 (2014).
- Harris, R. A., Trudell, J. R., Mihic, S. J. Ethanol's molecular targets. Science Signaling. 1 (28), (2008).
- Westerfield, M. The Zebrafish Book: A guide for the laboratory use of zebrafish Danio (Brachydanio) rerio. , University of Oregon Press. Eugene, OR. (1993).
- Lawson, N. D., Weinstein, B. M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Developmental Biology. 248 (2), 307-318 (2002).
- Hagedorn, M., Kleinhans, F. W., Artemov, D., Pilatus, U. Water Distribution and permeability of zebrafish embryos, Brachydanio rerio. Journal of Experimental Zoology. 278 (6), 356-371 (1997).
- Lippi, G., et al. The alcohol used for cleansing the venipuncture site does not jeopardize blood and plasma alcohol measurement with head-space gas chromatography and an enzymatic assay. Biochemia Medica. 27 (2), 398-403 (2017).
- Poklis, J. L., Wolf, C. E., Peace, M. R. Ethanol concentration in 56 refillable electronic cigarettes liquid formulations determined by headspace gas chromatography with flame ionization detector (HS-GC-FID). Drug Testing and Analysis. 9 (10), 1637-1640 (2017).
- Heit, C., et al. Quantification of Neural Ethanol and Acetaldehyde Using Headspace GC-MS. Alcoholism: Clinical and Experimental Research. 40 (9), 1825-1831 (2016).
- Chun, H. J., Poklis, J. L., Poklis, A., Wolf, C. E. Development and Validation of a Method for Alcohol Analysis in Brain Tissue by Headspace Gas Chromatography with Flame Ionization Detector. Journal of Analytical Toxicology. 40 (8), 653-658 (2016).
- Schlatter, J., Chiadmi, F., Gandon, V., Chariot, P. Simultaneous determination of methanol, acetaldehyde, acetone, and ethanol in human blood by gas chromatography with flame ionization detection. Human and Experimental Toxicology. 33 (1), 74-80 (2013).
- Schier, C. J., Mangieri, R. A., Dilly, G. A., Gonzales, R. A. Microdialysis of ethanol during operant ethanol self-administration and ethanol determination by gas chromatography. Journal of Visualized Experiments. (67), e4142(2012).
- Adalsteinsson, E., Sullivan, E. V., Mayer, D., Pfefferbaum, A. In vivo quantification of ethanol kinetics in rat brain. Neuropsychopharmacology. 31 (12), 2683-2691 (2006).
- Quertemont, E., Green, H. L., Grant, K. A. Brain ethanol concentrations and ethanol discrimination in rats: effects of dose and time. Psychopharmacology. 168 (3), 262-270 (2003).
- Flentke, G. R., Klinger, R. H., Tanguay, R. L., Carvan, M. J., Smith, S. M. An evolutionarily-conserved mechanism of calcium-dependent neurotoxicity. Alcoholism: Clinical and Experimental Research. 38 (5), 1255-1265 (2014).
- Reimers, M. J., Flockton, A. R., Tanguay, R. L. Ethanol- and acetaldehyde-mediated developmental toxicity in zebrafish. Neurotoxicology and Teratology. 26 (6), 769-781 (2004).
- Zhang, C., Ojiaku, P., Cole, G. J. Forebrain and hindbrain development in zebrafish is sensitive to ethanol exposure involving agrin, Fgf, and sonic hedgehog function. Birth Defects Research Part A: Clinical and Molecular Teratology. 97 (1), 8-27 (2013).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved