Method Article
Patient-Specific Polyvinyl Alcohol Phantom Fabrication with Ultrasound and X-Ray Contrast for Brain Tumor Surgery Planning
* These authors contributed equally
In This Article
Summary
This protocol describes the fabrication of a patient specific skull, brain and tumor phantom. It uses 3D printing to create molds, and polyvinyl alcohol (PVA-c) is used as the tissue mimicking material.
Abstract
Phantoms are essential tools for clinical training, surgical planning and the development of novel medical devices. However, it is challenging to create anatomically accurate head phantoms with realistic brain imaging properties because standard fabrication methods are not optimized to replicate any patient-specific anatomical detail and 3D printing materials are not optimized for imaging properties. In order to test and validate a novel navigation system for use during brain tumor surgery, an anatomically accurate phantom with realistic imaging and mechanical properties was required. Therefore, a phantom was developed using real patient data as input and 3D printing of molds to fabricate a patient-specific head phantom comprising the skull, brain and tumor with both ultrasound and X-ray contrast. The phantom also had mechanical properties that allowed the phantom tissue to be manipulated in a similar manner to how human brain tissue is handled during surgery. The phantom was successfully tested during a surgical simulation in a virtual operating room.
The phantom fabrication method uses commercially available materials and is easy to reproduce. The 3D printing files can be readily shared, and the technique can be adapted to encompass many different types of tumor.
Introduction
Phantoms mimicking the specific properties of biological tissues are a useful resource for various experimental and teaching applications. Tissue-mimicking phantoms are essential to characterize medical devices prior to their clinical use1,2 and anatomical phantoms are frequently used in the training of medical staff in all disciplines3,4,5,6,7. Patient-specific anatomical phantoms made with appropriate tissue-mimicking properties are often a critical part of the testing environment and can increase the confidence of clinicians who are learning to use a new device8. However, high manufacturing costs and complex fabrication processes often preclude the routine use of patient-specific phantoms. Here, a method is described for manufacturing a durable, patient-specific brain tumor model using readily available, commercial materials, which can be used for the training and validation of intraoperative ultrasound (US) using computerized tomography (CT) imaging. The phantom described in this study was created using data from a patient with a vestibular schwannoma (a benign brain tumor arising from one of the balance nerves connecting the brain and inner ear) who subsequently underwent surgery and tumor resection via a retrosigmoid suboccipital craniotomy10. The phantom was developed in order to test and validate an integrated intraoperative navigation system for use during this type of brain tumor surgery.
In order to be suitable for this application, the brain tumor phantom needs to possess several key properties. First, it should be made of non-toxic materials, so it can safely be used in a clinical training environment. Second, it should have realistic imaging properties; for the intended application, these specifically include ultrasound attenuation and CT contrast. Third, it should have similar mechanical properties to human tissue so that it can be handled in the same way. Fourth, the phantom should be based on real patient data, so that it is anatomically accurate and can be used for surgical planning and training. Finally, the materials used must be durable, so that the phantom can be used repeatedly.
In general, the tissue-mimicking material and fabrication method chosen for a phantom depends on the intended application. For rigid structures like the skull, the chosen property should not deform or be water-soluble and it should be able to maintain an accurate level of anatomical detail with repeated use; this is especially important when using the phantom for experiments where image registration is used and for surgical simulation purposes. Mineral oil based materials such as gel wax have been promising for ultrasound9,11,12 and photoacoustic13 imaging applications, however, when subjected to repeated mechanical deformation they become friable, so cannot withstand extended use, especially with standard microsurgical neurosurgery instruments. Agar and gelatin are aqueous materials that are also commonly used as tissue-mimicking materials. The additives needed to adjust the acoustic properties of these materials are well known14, but they have limited mechanical strength and are not particularly durable so are not suitable for this application, where the phantom needs to be repeatedly handled.
Polyvinyl alcohol cryogel (PVA-c) is a popular choice of tissue-mimicking material, because its acoustic and mechanical properties can easily be tuned by varying its freeze-thaw cycles. It has been shown that the properties of PVA-c are similar to those of soft tissues15,16,17,18. PVA-c based brain phantoms have been used successfully for ultrasound and CT imaging19. The material is strong enough to be used repeatedly, and it has a high degree of elasticity, so phantom tissue made of PVA-c can be manipulated without being permanently deformed. Polylactic acid (PLA) is a readily available rigid material and was used to manufacture the skull, however, a different printing material can be used in place of PLA, if it has similar mechanical properties and is not water soluble.
Brain phantoms in particular have been fabricated using different methods, depending on the level of complexity required and the tissues that need to be replicated20,21,22,23. Usually, a mold is used, and liquid tissue-mimicking material poured into it. Some studies have used commercial molds24 whilst others use 3D-printed custom molds of a healthy brain, and simulate brain lesions by implanting marker spheres and inflatable catheters19,25. To the best of the author’s knowledge, this is the first report of a 3D-printed patient-specific brain tumor phantom model created with tissue-mimicking ultrasound and X-ray properties. The total fabrication is visualized by the flowchart in Figure 1; the whole process takes around a week to complete.
Protocol
This study was conducted according to the principles expressed in the Declaration of Helsinki and was approved by the NHS Health Research Authority and Research Ethics Committee (18/LO/0266). Informed consent was obtained, and all imaging data were completely anonymized before analysis.
1. Data
- Obtain pre-operative contrast-enhanced T1-weighted Magnetic Resonance Imaging (MRI) and volumetric computed tomography (CT) data.
- If acquired in Digital Imaging and Communications in Medicine (DICOM) format, convert to Neuroimaging Informatics Technology Initiative26 (NiFTI) format for processing and analysis.
- Obtain intraoperative ultrasound data.
2. Segmentation
- Install software to segment the patient data with.
- Skull segmentation
NOTE: The steps involved in segmenting the skull broadly follow those outlined by Cramer and Quigley27 on https://radmodules.com/, but are adapted to create an appropriately-sized craniotomy.- Load the patient’s volumetric CT scan in segmentation software, open the Segment Editor module and create new segmentation named ‘Skull’.
- Use the ‘Threshold’ function to highlight the skull.
- Remove any unwanted segmentations (e.g., skin calcifications, mandible, C1/2, styloid process, the CT patient frame, and any annotations embedded within the image). Use the ‘Scissors’ function to remove parts when viewing the model in 3D and make use of the ‘Islands’ function after manually disconnecting any unwanted structures using the ‘Erase’ function.
- Manually correct any gaps in the segmentation that were missed during thresholding using the ‘Paint’ and ‘Draw’ functions (e.g., lamina papyracea, cortical edge of the mastoid bone and ethmoid bone).
- Use the ‘Paint’ and ‘Draw’ functions to fill in the foramen magnum and create a 5 mm protruding spike upon which the lower part of phantom model can be secured.
NOTE: The location of the spike is best determined on the coronal and sagittal image planes. - Apply the ‘Smoothing’ function. Use a median smoothing setting of 1.0 mm (3 × 3 × 1 pixels) to minimize the amount of detail lost.
NOTE: If the phantom model must include a complete intact skull (e.g., to facilitate surgical simulation of creating an appropriately located craniotomy), move to step 2.2.15; however, if a craniotomy is required in the model, complete steps 2.2.7 to 2.2.14. - Click ‘Add’ to add a new segmentation and name it ‘Skull Craniotomy’.
- In the ‘Segmentations’ module, copy the ‘Skull’ segmentation across to ‘Skull Craniotomy’ using the ‘Copy/Move Segments’ tab.
NOTE: Both the ‘Skull’ and ‘Skull Craniotomy’ segmentations are needed in order to be able to perform the functions described in steps 2.2.9 to 2.2.13 - Use the ‘Scissors’ function to remove an appropriately-sized craniotomy in ‘Skull Craniotomy’.
NOTE: Creating the craniotomy this way will, also, remove an addition portion of skull on the opposite side hence the need for steps 2.2.11 to 2.2.14. - Click ‘Add’ and add a new segmentation; name it ‘Craniotomy Only’.
- In ‘Craniotomy Only’ select the segmentation ‘Skull Craniotomy’ and use the ‘Logical Operator’ function to subtract ‘Skull Craniotomy’ from ‘Skull’.
- Use the ‘Scissors’ function to erase everything except the desired craniotomy on the correct side of the tumor, saving ‘Craniotomy Only’.
- In ‘Skull Craniotomy’ use the ‘Logical Operator’ function to subtract ‘Craniotomy only’ from ‘Skull’ and save.
- Open ‘Segmentations’ module and export the ‘Skull Craniotomy’ as a stereolithography (STL) file.
- Open 3D modeling software and import the STL file ‘Skull Craniotomy’.
NOTE: If the model appears in striped pink complete the ‘Flip Normals’ function by selecting the complete model (Select | Double click) and then ‘Edit | Flip Normals’. The model will now turn grey and can be edited. Ensure ‘View Objects Browser’ is turned on. - Reduce the number of triangles to improve the computational time.
- Select the complete model (Select | Double click turns the model orange) then ‘Edit | Reduce’. The default ‘Reduce’ function is set at 50% so repeat until the desired reduction is achieved. Aim for a total number of triangles < 500,000.
- Apply ‘Smoothing’ function ensuring the ‘Shape Preserving’ box remains ticked. Select the complete model then ‘Deform | Smooth’.
- Click ‘Analysis’ then ‘Inspector’ and use this function to detect any small defects in the model and click auto-repair (suggest ‘Flat-fill’ selection).
- Cut 'Skull' to create a top and bottom using the ‘Edit/Plane’ cut function. Select ‘Keep Both Slices’ and ‘Remeshed’ fill type. Change skull to transparent with ‘Shaders’ function to provide a better internal view of the skull and adjust the plane so that it is parallel to the skull base.
- Separate shells by selecting ‘Edit | Separate shells’ and rename ‘Skull_Top’ and ‘Skull_Bottom’ within the objects browser.
NOTE: Do not move their positions. Click the eye icon to remove one or the other from view. - Click ‘Meshmix’ then select ‘Cylinder’ to create a dowel and edit size to 4 mm × 10 mm × 4 mm (‘Edit | Transform’). Hide ‘Skull_Bottom’ by clicking the eye icon to remove from view.
- Select ‘Edit | Align’ planes. An additional transparent cylinder will appear. In the ‘Align’ window, choose ‘Surface point’ (left click end transparent cylinder) for the ‘Source’ and ‘Surface point’ (Shift + left click undersurface of ‘Skull_Top’) for the ‘Destination.’
- Using the ‘Edit | Transform’ function move dowel into skull using the green arrow and adjust position with blue and red arrows. Rename ‘Dowel_Anterior’.
- In the object browser make 3 copies and rename ‘Dowel_Posterior’, ‘Dowel_Left’ and ‘Dowel_Right’.
- Move each dowel to the desired location using the ‘Edit | Transform’ function.
NOTE: Do not move or change the position of the dowel in the green plane. - Create copies of each but keep all copies in the same location and create an additional dowel and resize to 3 mm × 10 mm × 3 mm. Rename ‘Dowel’.
- Create holes for Dowels in the skull using the ‘Boolean Difference’ function. Select ‘Skull_Top’ first and then select a dowel in the object browser. In the ‘Boolean Difference’ tab ensure ‘Auto-reduce’ is switch off. Repeat for each dowel in turn.
- Hide ‘Skull_Top’ and view ‘Skull_Bottom’ repeating the above ‘Boolean Difference’ function for each dowel in turn.
- Export ‘Skull_Top’, ‘Skull_Bottom’ and ‘Dowel’ as separate binary STL files.
- Brain tissue segmentation
- Upload the contrast enhanced T1 MRI of the brain to http://niftyweb.cs.ucl.ac.uk/program.php?p=GIF and download its output. This is an open-source parcellation tool for T1-weighted images that utilizes a Geodesic Information Flow (GIF) algorithm28 to perform brain extraction and tissue segmentation.
- Open segmentation software and load the contrast enhanced T1 MRI and GIF parcellation output file.
- Open the ‘Segment Editor’ module and create a new segmentation.
- Select the appropriate labels and combine them to form a single segmentation. For example, cerebral and diencephalon label maps can be combined to create one model, referred to as ‘Brain’ and midbrain, brainstem, cerebellum and vermian structures can be combined to create a second model referred to as ‘Cerebellum’.
- Use the ‘Smoothing’ function (suggested median 2.00 mm, 5 × 5 × 3 pixels).
- Use the ‘Scissors’ function to remove any unwanted or erroneous segmentations.
- Save ‘Brain’ and ‘Cerebellum’ segmentations.
- Open ‘Segmentations’ module and export ‘Brain’ and ‘Cerebellum’ as STL files.
- Tumor segmentation
- Open segmentation software and load the contrast enhanced T1 MRI.
- Open the ‘Segment Editor’ module and create new segmentation named ‘Tumor’.
- Use the ‘Threshold’ function to highlight the tumor.
- Correct the segmentation using the ‘Paint’, ‘Draw’ and ‘Erase’ functions.
- Apply the ‘Smoothing’ function (suggested median 2.00 mm 5 x 5 x 3 pixels).
- Create a new segmentation named ‘Cerebellum_Tumor’.
- Combine the ‘Cerebellum’ model and ‘Tumor’ using the ‘Logical Operators | Add’ function.
- Save ‘Tumor’ and ‘Cerebellum_Tumor’ segmentations.
- Open ‘Segmentations’ module and export ‘Tumor’ and ‘Cerebellum_Tumor’ as STL files.
NOTE: At the end of the segmentation process, the following files are available: ‘Skull_Top’, ‘Skull_Bottom’, ‘Dowel’, ‘Brain’, ‘Cerebellum’, ‘Tumor’, ‘Cerebellum_Tumor’.
3. 3D Printing of Brain/Tumor Molds and Skull
- Create the brain and tumor molds
- Split the ‘Brain’ segmentation into two hemispheres, using the ‘Plane cut’ tool in 3D modeling software.
- Save each hemisphere as a separate STL file ‘Brain right’ and ‘Brain left’.
- Import the STL file ‘Tumor’ into computer-aided design (CAD) software.
- Click the 'mesh' tab and then use the ‘Reduce’ function to reduce the size of the model so that it can be handled by the program – the aim is to reduce the size as much as possible, whilst still retaining all the detail necessary.
- Click the 'solid' tab and use the ‘Mesh to BRep’ tool to convert the imported mesh to a body that can be manipulated. If this action cannot be completed, the mesh was not reduced enough in step 3.1.3.
- Click ‘Create’ then ‘Box’ and draw a box around the tumor. Select to create this as a ‘New Body’ and rotate the view to ensure the box completely encloses the tumor on all sides.
- In the modify tab, use the ‘Combine’ tool to cut the tumor (the ‘Tool Body’) from the box (the ‘Target Body’). This will then leave a box with a hollow shape of the tumor inside it.
- Check that the hollowed-out box is present. Cut this box into an appropriate number of pieces so that once the mold is filled, it can be prized apart without damaging the phantom inside. For the tumor here, it is enough to split the box in two, but for the other parts of the phantom, more pieces are needed.
- Create planes through the box in the places that the mold needs to be cut. Click ‘Construct’ then ‘Midplane’ to create a plane through the center of the box. Right click on the created plane and choose ‘Offset Plane’ to position the plane more precisely.
- Use the ‘Split Body’ function in the ‘Modify’ tab to split the mold along the planes created.
- Move the individual pieces of the mold, by right clicking and selecting ‘Move/Copy’, so that all the pieces are facing outwards.
- Add rivets to the faces of each piece of the mold (so it can fit together securely), by clicking ‘Create sketch’ then ‘Centre diameter circle’ and on each face, drawing small circles. Right click then ‘Extrude’ these circles outwards a few millimeters on one face and extrude them inwards on the corresponding face.
NOTE: The circles that are extruded inwards need to be slightly bigger - approximately 1.5 mm - than those that are extruded outwards, so that they will fit together snugly. - Save each piece of the mold as a separate STL file.
- Repeat steps 3.1.4 – 3.1.14 for ‘Brain left’, ‘Brain right’ and ‘Cerebellum tumor’.
NOTE: Using the file ‘Cerebellum tumor’ rather than just ‘Cerebellum’ to create the mold means that the mold will have a space in it for the tumor to be inserted during construction.
- Print the 3D molds
- Install or open 3D printing software.
- Open the STL file for each piece of the mold in the printing software and rotate it so that it lies flat against the build plate. It is possible to add multiple mold pieces to the build plate and to print these simultaneously.
- Choose a large layer height (around 0.2 mm) and low infill value (around 20%) for faster printing. Print the molds using a rigid material such as Polylactic acid (PLA). If the molds are positioned appropriately, support material is not necessary.
- Print the Skull
- Open the ‘Skull Top’ file in the printing software and choose a large layer height (around 0.2 mm) and low infill value (around 20%).
- Print the skull model in PLA but in contrast to step 3.2.3, support material will be required, so select to ‘Add support’ in the software. PVA is used as the support material as it can later be dissolved away with water.
- Repeat steps 3.3.1 and 3.3.2 for ‘Skull Bottom’.
- Once the top and bottom of the skull have been printed, submerge them in water overnight to dissolve away the PVA support material.
NOTE: The support material will dissolve away much faster if warm water is used, but if the water is too warm, it will deform the printed PLA. Therefore, it is preferable to use cool water and leave the print submerged overnight.
4. Preparation of PVA-c
- Measure 200 g of PVA powder and set to the side.
- Heat 1800 g of deionized water to 90 °C and add to a 2L conical flask.
NOTE: The water needs to be almost boiling so the PVA powder will dissolve readily, but if the water reaches 100 °C, some will be lost to evaporation, which is to be avoided. - Suspend the conical flask in a temperature-controlled water bath set at 90 °C.
- Position an electronic stirrer in the flask, ensuring it does not touch the bottom or sides, and set the speed to 1500 rpm.
NOTE: Check that the water is stirring evenly and there are not stagnant points at the sides or bottom. - Gradually add the PVA powder to the conical flask, over around 30 min, then leave it to stir for around another 90 min. The resulting gel is the tissue-mimicking material PVA-c.
- Remove conical flask from the water bath and pour the contents into a beaker. Cover the top with cling film to prevent the formation of a skin on top of the PVA-c. Leave the PVA-c to cool to room temperature (around 20 °C). Once cooled, the PVA-c will be transparent. Tiny white crystals may be seen in the PVA-c, but any bubbles appearing on the surface must be gently scraped off.
- Add 0.5 w/w% potassium sorbate to the PVA-c as a preservative, and manually stir well.
- The PVA-c can be left at room temperature if covered in cling film for a few days before it is poured into molds.
5. Phantom Assembly
- Measure out enough PVA-c to fill the tumor mold into a beaker.
- To the PVA-c for the tumor, add 1 w/w% glass microspheres for ultrasound contrast and 5 w/w% Barium Sulfate for X-ray contrast, and stir by hand.
NOTE: It may be necessary to measure out excess PVA-c for the tumor so that these percentages are a measurable amount. - Sonicate the beaker to ensure homogenous mixing of the additives.
- Leave to cool and allow any bubbles formed to escape, around 10 min, then scrape any bubbles from the surface.
NOTE: Do not leave for extended period once the glass spheres have been added, no longer than around 10 min, before pouring the PVA-c into a mold, as the glass spheres will settle to the bottom of the beaker. Once the phantom has been frozen, this will no longer be a concern, and the final phantom can be used at room temperature. - Secure the tumor mold together (tape can be used to cover the joins in the mold) and pour in the PVA-c through the hole in the top of the mold. Leave for a few minutes to allow any bubbles formed in the pouring process to escape through the hole, then place straight into the freezer.
- Perform two freeze-thaw cycles on the tumor; each cycle here consists of 6 h of freezing at -20 °C and 6 h of thawing at room temperature. Then, carefully remove from mold.
- Place the tumor into the corresponding space for it in the cerebellum mold, then construct the rest of the cerebellum mold and secure it together.
- To the remaining PVA-c add 0.05 w/w% glass microspheres, then repeat steps 5.1.3 and 5.1.4.
- Pour the PVA-c into the cerebellum mold, allowing it to surround the tumor that has been placed inside. Additionally, pour the mixture into the molds for each brain hemisphere.
- Perform two freeze-thaw cycles on each brain hemisphere and the cerebellum; each cycle here consists of 24 h of freezing at -20 °C and 24 h of thawing at room temperature.
NOTE: Cycles with 12 h freezing followed by 12 h thawing also effective, to allow the phantom to be created in less time. 24 h was chosen for ease of application, to avoid returning to the lab every 12 h. - Once the phantoms have thawed for the second time, carefully remove them from the molds and place into the printed skull.
NOTE: When not in use, the completed PVA-c phantoms should be stored in an airtight container in the fridge, and can be kept for a few weeks in this way - For completion, place the ‘Cerebellum tumor’ phantom on the spike at the base of the ‘Skull Bottom’ model. The models of two brain hemispheres (‘Brain left’ and ‘Brain right’) are placed on top and slot into the uppermost part of the ‘Cerebellum tumor’.
- Place the four dowels in each space on the ‘Skull Bottom’ model and place ‘Skull Top’ model on top. If required, the model may then be maneuvered into the desired position to simulate intraoperative use in surgery.
6. Phantom Imaging
- Ultrasound Imaging
- Apply ultrasound gel to the imaging probe.
NOTE: Gel is not used intraoperatively but may be used in simulation and does not significantly change the clinical workflow or the quality of the acquired images. - Image the brain and tumor through the craniotomy, with a clinical scanner and burr hole probe.
- Apply ultrasound gel to the imaging probe.
- CT Imaging
- Image the whole phantom in a CT scanner.
Results
Following the described protocol, an anatomically realistic phantom was fabricated, which consists of a patient-specific skull, brain and tumor. The relevant anatomical structures for the phantom (skull, brain, tumor) are segmented using patient MRI and CT data (Figure 2a,b). The patient intra-operative ultrasound data (Figure 2c; Figure 2d shows the same image as Figure 2c, but with the tumor outlined) was used to compare the phantom images to the real patient images.
Meshes were created for each piece of the model (Figure 3), and these were then used to manufacture the 3D molds. The molds were easily printed on a commercial printer and assembled by slotting the pieces together. The cerebellum mold was the most complex to design and assemble (Figure 4). The skull (Figure 5a) was the most difficult part to print as it required support material, so was a slow process; the whole print took a total of three days to complete, which is a limiting factor in the protocol.
The completed phantom (Figure 5) was a realistic model of a patient skull, brain and tumor. The two brain hemispheres (Figure 5b) were produced separately, and have a realistic appearance, featuring the gyri and sulci of the brain. The whole phantom is white in color, as this is the natural color of PVA-c; this can easily be changed by adding dye but was not necessary for the application. The cerebellum (Figure 5c) fits comfortably into the base of the printed skull and the brain hemispheres sit on top of this. The tumor is easily visible in the cerebellum, as the extra contrast added to the tumor results in it being an off-white color that separates it from the surrounding material, which is it securely attached to.
The phantom was imaged with both CT and ultrasound (Figure 6a,b). Barium sulfate was used to give the tumor appropriate CT contrast, and the phantom image (Figure 6a) shows that this was achieved, as the tumor is clearly visualized. The skull was not printed with 100% infill, in order to reduce the time taken for printing. Therefore, the skull does not look entirely realistic in the CT images, because the lattice structure of the print can be seen. This is not a problem for the application, as only the outline of the skull is needed for the neuronavigation system. The skull could be printed with 100% infill to avoid this reduced accuracy of the CT image, but would add time onto the printing process. Glass microspheres were added to the cerebellum, brain hemispheres and tumor for ultrasound contrast. The results show that the tumor is also visible with ultrasound imaging (Figure 6b) and can be distinguished from the surrounding tissue. On visual inspection, the ultrasound images obtained from the phantom (Figure 6b), and those obtained from the patient (Figure 2c) show that the contrast agents used in the phantom were effective for creating realistic imaging properties.
The phantom was tested during surgical simulation in a virtual operating room (Figure 7). The phantom model was positioned on the surgical operating table using a standard skull clamp and the CT scan of the phantom was registered using a clinical neuronavigation system. A retrosigmoid approach to the tumor was simulated and the tumor was imaged using a clinical ultrasound system with a burr hole ultrasound transducer. During the surgical simulation, the phantom model proved to be stable and no damage was observed from manipulating the phantom in the same way the human brain would be during this procedure, so it could be used repeatedly under the same conditions.
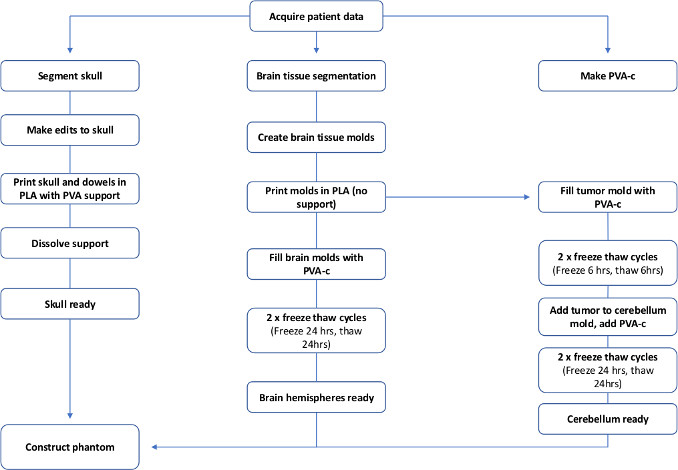
Figure 1: Flowchart to show the steps required to make a patient specific PVA-c brain phantom. Please click here to view a larger version of this figure.
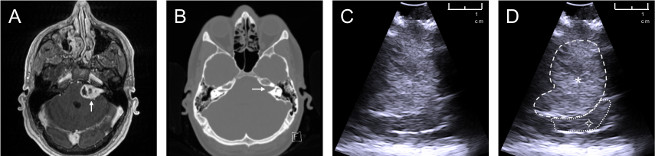
Figure 2: Patient data used to create phantom model. Data sources of a patient with a left sided vestibular schwannoma: (a) axial contrast-enhanced T1-weighted MRI, white arrow pointing towards tumor; (b) axial non-contrast CT scan windowed to highlight bone, white arrow pointing towards an expanded internal auditory meatus caused by the tumor; (c) intraoperative ultrasound image obtained during vestibular schwannoma surgery; (d) annotated intraoperative ultrasound image ![]() : tumor (hyperechoic on ultrasound),
: tumor (hyperechoic on ultrasound), ![]() : brain (cerebellum). Please click here to view a larger version of this figure.
: brain (cerebellum). Please click here to view a larger version of this figure.
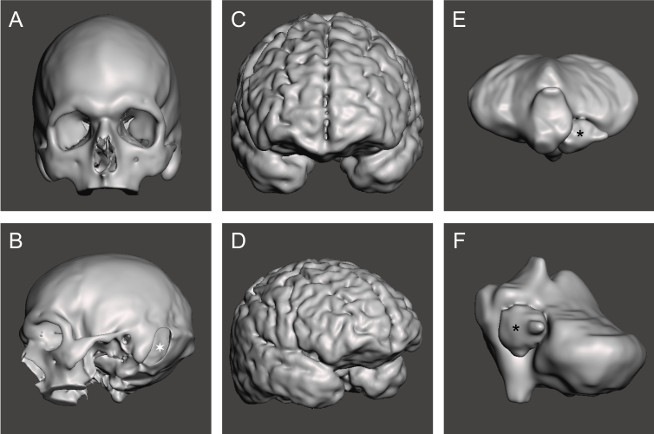
Figure 3: Completed meshes for each section of the phantom. STL mesh for (a,b) skull, ![]() : left sided retrosigmoid craniotomy; (c,d) cerebral hemispheres; (e,f) tumor and cerebellum,
: left sided retrosigmoid craniotomy; (c,d) cerebral hemispheres; (e,f) tumor and cerebellum, ![]() : tumor. Please click here to view a larger version of this figure.
: tumor. Please click here to view a larger version of this figure.
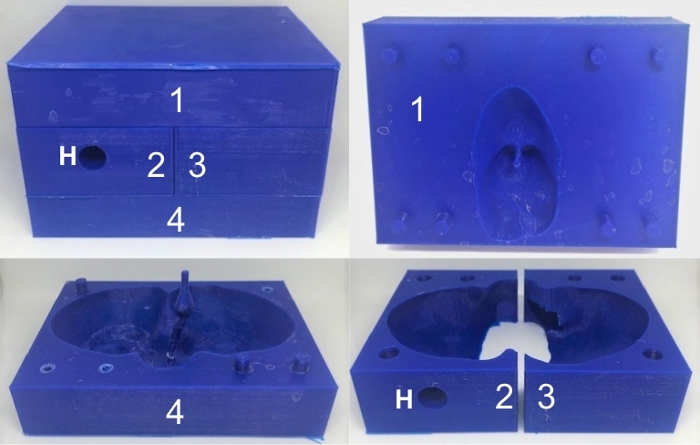
Figure 4: 3D printed cerebellum mold. 3D printed cerebellum mold fully constructed (top left) and the separate pieces, which are numbered from 1 to 4. The hole in piece 2 (denoted by ‘H’) enables the PVA-c to be poured into the mold. Please click here to view a larger version of this figure.
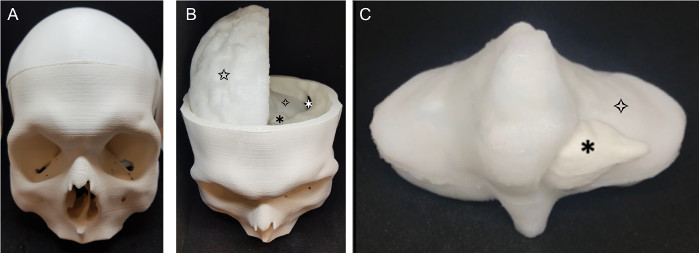
Figure 5: Completed phantom. The finished phantom (a) skull (b) phantom with skull top removed: ![]() : retrosigmoid craniotomy,
: retrosigmoid craniotomy, ![]() : tumor,
: tumor, ![]() brain (cerebellum),
brain (cerebellum), ![]() brain (right cerebral hemisphere); (c) cerebellum and tumor:
brain (right cerebral hemisphere); (c) cerebellum and tumor: ![]() : tumor,
: tumor, ![]() brain (cerebellum). Please click here to view a larger version of this figure.
brain (cerebellum). Please click here to view a larger version of this figure.
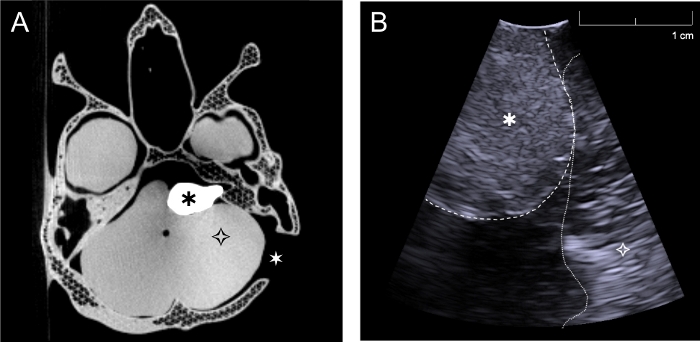
Figure 6: CT and ultrasound images acquired with the phantom. (a) Axial CT image of complete phantom through the level of the skull base and tumor, (b) Intraoperative ultrasound image of phantom acquired with burr hole ultrasound probe through the retrosigmoid craniotomy in a plane approximately perpendicular to the skull (Simulating surgery, the cerebellum was retracted slightly in order to image directly on the tumor). ![]() : tumor,
: tumor, ![]() brain (cerebellum),
brain (cerebellum), ![]() : left sided retrosigmoid craniotomy. Please click here to view a larger version of this figure.
: left sided retrosigmoid craniotomy. Please click here to view a larger version of this figure.
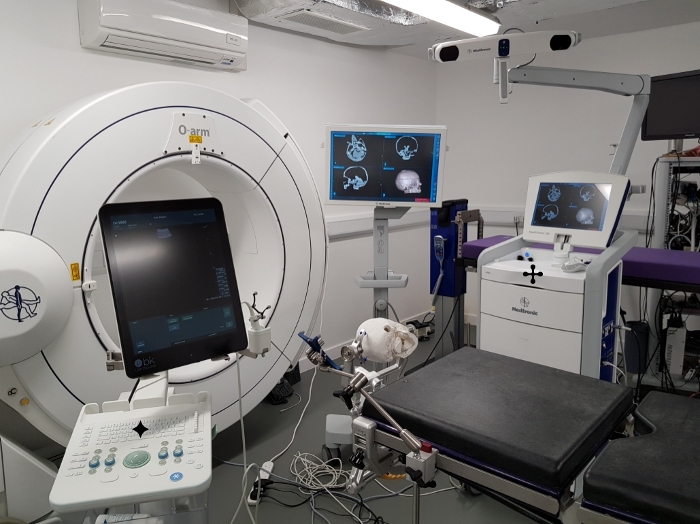
Figure 7: Testing the phantom during surgical simulation. Testing the phantom model through surgical simulation in a virtual operating room. ![]() : neuronavigation system displaying the registered scan of the CT phantom model,
: neuronavigation system displaying the registered scan of the CT phantom model, ![]() : ultrasound system used to image the phantom with a burr hole ultrasound transducer (seen positioned next to the ultrasound monitor). Note the model pictured here is based on data acquired from different patient with a right sided tumor. Please click here to view a larger version of this figure.
: ultrasound system used to image the phantom with a burr hole ultrasound transducer (seen positioned next to the ultrasound monitor). Note the model pictured here is based on data acquired from different patient with a right sided tumor. Please click here to view a larger version of this figure.
Discussion
This protocol details the fabrication process of a patient specific brain phantom, which includes the skull, brain, and vestibular schwannoma tumor. 3D printing methods allowed for anatomically accurate detail to be achieved. The phantom described here was successfully manufactured with the desired level of anatomical detail; CT and ultrasound imaging were used to demonstrate that the tumor was easily visualized with both modalities. The tissue mimicking material, PVA-c, is well established as a tissue-mimicking material for ultrasonic phantoms; its acoustic and mechanical properties can be tuned with additives and the number of freeze-thaw cycles. The material is readily available, simple to use and non-toxic. With repeated use, the phantom had sufficient durability to withstand manipulation and contact with an ultrasound probe during physical simulations of vestibular schwannoma surgery.
Several key steps were identified as being critical to the fabrication process. First, the segmentation of structures for inclusion in the phantom must include the desired level of anatomical detail. The creation of accurate STL files and 3D molds then follows naturally. Secondly, the positioning of planes within the cerebellum mold in step 3.1.9 must be considered carefully, so that the phantom can be readily removed, without damage; it must be cut into enough pieces to allow anatomical details to be retained, whilst enabling the phantom to be removed without getting stuck in the mold. In this case, several iterations were tested and finally the mold was cut into four separate pieces. The third key consideration is that during the PVA-c manufacturing process (section 4), the PVA-c must be left to cool to room temperature (step 4.1.6). If this step is missed and hot PVA-c is added to the molds, it can cause the molds to melt or distort. It is also crucial that once the glass spheres are added (steps 5.1.2 – 5.1.4), the PVA-c is not left to sit for more than around 10 minutes; if left for a prolonged period of time, the glass spheres will settle to the bottom, and the resulting phantom will have inhomogeneous ultrasound contrast29. Once the glass spheres are added, the PVA-c must be added directly into the molds and placed into the freezer. After the first freeze cycle, the glass spheres will be secured in the place, and the phantom can be used at room temperature. Finally, it is important that the molds are carefully sealed (e.g., with tape) before the PVA-c is added, to minimize leakage of the mixture through gaps where the separate pieced of the mold joined together.
The protocol has several limitations. For example, some specialist equipment is required, including a water bath and an electronic stirrer. A sonicator is also used as part of this protocol, but the sonication step (5.1.3) could be replaced with additional electronic stirring; however, with this alternative, it would take longer to achieve a homogeneous mixture than is possible with the use of sonication. One limitation of PVA-c is that it degrades over time and becomes moldy. The addition of potassium sorbate, as described here, increases the phantom’s shelf-life, although it must still be kept in an air-tight container. A second limitation of PVA-c is that freeze-thaw cycles are required, which increases the amount of time required to make a phantom. To minimize phantom fabrication time, a key consideration is the speed of freezing and thawing; once the phantom is either fully frozen or fully thawed, the time that it remains in that state does not significantly affect the final phantom16,30. Therefore, the cycle lengths used can be varied, provided that the phantom is fully frozen and thawed at each stage in the cycle. For instance, the tumor in the phantom of this study is very small, so shorter cycles could be used for the tumor than for the brain. Finally, 3D printing the molds and skull is a time-consuming process which consumes a significant portion (3 days) of the total time (1 week) required to fabricate a phantom with this protocol. The printer used was a commercial model from 2018; the printing process could be completed in shorter time frames with the use of newer, faster printers.
The brain phantom presented here could be used directly for clinical training and validation of neuronavigation systems. As the tissue mimicking material, PVA-c enables the resulting phantom to be used repeatedly, for example as a training tool or for the validation of intraoperative ultrasound in vestibular schwannoma surgery, as it is a durable and non-toxic material. As such, the fabrication method is complementary to those previously described in which 3D printing was used to create patient specific brain phantoms20,21,22,23,24,25. The use of PVA-c as the TMM makes the phantom suitable for use in simulation of neurosurgery, as the material can withstand repeated manual manipulation and contact from an ultrasound probe. This work sets the stage for further quantitative validation studies. The phantom method described here is very versatile and could be used to fabricate many types of patient-specific tumor phantoms, extending from the brain to other organs, with compatibility across several imaging modalities.
Disclosures
The authors do not have any conflicts of interest to declare.
Acknowledgements
The authors thank Daniil Nikitichev and Steffi Mendes for their advice on using Meshmixer and Fernando Perez-Garcia for his advice on using 3D Slicer and for providing us code to automate some of the processing steps.
This work was supported by Wellcome Trust [203145Z/16/Z; 203148/Z/16/Z; WT106882], EPSRC [NS/A000050/1; NS/A000049/1], MRC [MC_PC_17180] and National Brain Appeal [NBA/NSG/SBS] funding. TV is supported by a Medtronic Inc / Royal Academy of Engineering Research Chair [RCSRF1819\7\34].
Materials
| Name | Company | Catalog Number | Comments |
| AutodeskFusion 360 | Autodesk Inc., San Rafael, California, United States | https://www.autodesk.co.uk/products/fusion-360/overview | CAD software |
| Barium sulphate | Source Chemicals | - | |
| CT scanner | Medtronic Inc, Minneapolis, USA | - | O-arm 3D mobile X-ray imaging system |
| Glass microspheres | Boud Minerals | ||
| Mechanical stirrer | IKA | 4442002 | Eurostar Digital 20, IKA |
| Meshmixer | Autodesk Inc., San Rafael, California, United States | http://www.meshmixer.com | 3D modelling software. Version 3.5.484 used |
| Neuronavigation system | Medtronic Inc, Minneapolis, USA | - | S7 Stealth Station |
| PLA | Ultimaker (Ultimaker BV, Utrecht, Netherlands) | UM9016 | |
| Potassium sorbate | Meridianstar | - | |
| PVA | Ultimaker | - | |
| PVA powder | Sigma-Aldrich | 363146 | 99%+ hydrolysed, average molecular weight 85,000-140,000 |
| Sonicator | Fisher Scientific | 12893543 | |
| Ultimaker Cura | Ultimaker BV, Utrecht, Netherlands | https://ultimaker.com/software/ultimaker-cura | 3D printing software. Version 4.0.0 used |
| Ultimaker S5 Printer | Ultimaker BV, Utrecht, Netherlands | - | |
| Ultrasound scanner | BK Medical, Luton, UK | - | BK 5000 scanner |
| Water bath | IKA | 20009381 | HBR4 control, IKA |
| 3D Slicer | http://slicer.org | - | Software used to segment patient data. Version 4.10.2 used |
References
- Culjat, M. O., Goldenberg, D., Tewari, P., Singh, R. S. A review of tissue substitutes for ultrasound imaging. Ultrasound in Medicine and Biology. 36 (6), 861-873 (2010).
- Hwang, J., Ramella-Roman, J. C., Nordstrom, R. Introduction: Feature Issue on Phantoms for the Performance Evaluation and Validation of Optical Medical Imaging Devices. Biomedical Optics Express. 3 (6), 1399 (2012).
- Maul, H., et al. Ultrasound simulators: Experience with the SonoTrainer and comparative review of other training systems. Ultrasound in Obstetrics and Gynecology. 24 (5), 581-585 (2004).
- Craven, C., et al. Development of a modelled anatomical replica for training young neurosurgeons. British Journal of Neurosurgery. 28 (6), 707-712 (2014).
- Zhang, L., Kamaly, I., Luthra, P., Whitfield, P. Simulation in neurosurgical training: a blueprint and national approach to implementation for initial years trainees. British Journal of Neurosurgery. 30 (5), 577-581 (2016).
- Leff, D. R., et al. Validation of an oncoplastic breast simulator for assessment of technical skills in wide local excision. British Journal of Surgery. 103 (3), 207-217 (2016).
- Hunt, A., et al. Low cost anatomically realistic renal biopsy phantoms for interventional radiology trainees. European Journal of Radiology. 82 (4), 594-600 (2013).
- Pacioni, A., et al. Patient-specific ultrasound liver phantom: materials and fabrication method. International Journal of Computer Assisted Radiology and Surgery. 10 (7), 1065-1075 (2015).
- Maneas, E., et al. Anatomically realistic ultrasound phantoms using gel wax with 3D printed moulds. Physics in Medicine and Biology. 63 (1), (2018).
- Samii, M., Matthies, C. Management of 1000 vestibular schwannomas (acoustic neuromas): hearing function in 1000 tumor resections. Neurosurgery. 40 (2), 242-248 (1997).
- Cabrelli, L. C., Pelissari, P. I. B. G. B., Deana, A. M., Carneiro, A. A. O., Pavan, T. Z. Stable phantom materials for ultrasound and optical imaging. Physics in Medicine and Biology. 62 (2), 432-447 (2017).
- Vieira, S. L., Pavan, T. Z., Junior, J. E., Carneiro, A. A. O. Paraffin-Gel Tissue-Mimicking Material for Ultrasound-Guided Needle Biopsy Phantom. Ultrasound in Medicine & Biology. 39 (12), 2477-2484 (2013).
- Maneas, E., et al. Gel wax-based tissue-mimicking phantoms for multispectral photoacoustic imaging. Biomedical Optics Express. 9 (3), 1151 (2018).
- Madsen, E. L., Hobson, M. A., Shi, H., Varghese, T., Frank, G. R. Tissue-mimicking agar/gelatin materials for use in heterogeneous elastography phantoms. Physics in Medicine and Biology. 50 (23), 5597-5618 (2005).
- Duboeuf, F., et al. Investigation of PVA cryogel Young’s modulus stability with time, controlled by a simple reliable technique. Medical Physics. 36 (2), 656-661 (2009).
- Fromageau, J., Brusseau, E., Vray, D., Gimenez, G., Delachartre, P. Characterization of PVA cryogel for intravascular ultrasound elasticity imaging. IEEE Transactions on Ultrasonics, Ferroelectrics and Frequency Control. 50 (10), 1318-1324 (2003).
- Fromageau, J., et al. Estimation of polyvinyl alcohol cryogel mechanical properties with four ultrasound elastography methods and comparison with gold standard testings. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. 54 (3), 498-508 (2007).
- Khaled, W., et al. Evaluation of Material Parameters of PVA Phantoms for Reconstructive Ultrasound Elastography. 2007 IEEE Ultrasonics Symposium Proceedings. , 1329-1332 (2007).
- Chen, S. J. S., et al. An anthropomorphic polyvinyl alcohol brain phantom based on Colin27 for use in multimodal imaging. Medical Physics. 39 (1), 554-561 (2012).
- Ploch, C. C., Mansi, C. S. S. A., Jayamohan, J., Kuhl, E. Using 3D Printing to Create Personalized Brain Models for Neurosurgical Training and Preoperative Planning. World Neurosurgery. 90, 668-674 (2016).
- Weinstock, P., et al. Creation of a novel simulator for minimally invasive neurosurgery: Fusion of 3D printing and special effects. Journal of Neurosurgery: Pediatrics. 20 (1), 1-9 (2017).
- Grillo, F. W., et al. Patient-specific neurosurgical phantom: assessment of visual quality, accuracy, and scaling effects. 3D Printing in Medicine. 4 (1), (2018).
- Tsai, A., et al. Creation and Validation of a Simulator for Neonatal Brain Ultrasonography: A Pilot Study. Academic Radiology. 24 (1), 76-83 (2017).
- Reinertsen, I., Collins, D. L. A realistic phantom for brain-shift simulations. Medical Physics. 33 (9), 3234-3240 (2006).
- Mobashsher, A. T., Abbosh, A. M., Wang, Y. Microwave system to detect traumatic brain injuries using compact unidirectional antenna and wideband transceiver with verification on realistic head phantom. IEEE Transactions on Microwave Theory and Techniques. 62 (9), 1826-1836 (2014).
- Cox, R. W., et al. A (sort of) new image data format standard: NiFTI-1. 10th Annual Meeting of the Organization for Human Brain Mapping. 22, (2004).
- Cramer, J., Quigley, E. Advanced Visualization and 3D Printing. Learning Lab at the Society for Imaging Informatics in Medicine annual meeting. , (2019).
- Cardoso, M. J., et al. Geodesic Information Flows: Spatially-Variant Graphs and Their Application to Segmentation and Fusion. IEEE Transactions on Medical Imaging. 34 (9), 1976-1988 (2015).
- Dong, J., Zhang, Y., Wei-Ning, L. Walled vessel-mimicking phantom for ultrasound imaging using 3D printing with a water-soluble filament: design principle, fluid-structure interaction (FSI) simulation, and experimental validation. Physics in Medicine & Biology. , 0 (2020).
- Jiang, S., Liu, S., Feng, W. PVA hydrogel properties for biomedical application. Journal of the Mechanical Behavior of Biomedical Materials. 4 (7), 1228-1233 (2011).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved