Method Article
Measuring Caenorhabditis elegans Sensitivity to the Acetylcholine Receptor Agonist Levamisole
In This Article
Summary
The present protocol describes an assay to determine response to levamisole, a pharmacological agonist of one class of Caenorhabditis elegans acetylcholine receptors. In this liquid levamisole swim assay, researchers visually observe and quantitate the time-dependent paralysis of animals cultivated in 24-well plates.
Abstract
At the neuromuscular junction (NMJ), the binding of the excitatory neurotransmitter acetylcholine (ACh) to postsynaptic receptors leads to muscle contraction. As in vertebrate skeletal muscle, cholinergic signaling in the body wall muscles of the model organism Caenorhabditis elegans is required for locomotion. Exposure to levamisole, a pharmacological agonist of one class of ACh receptors on the body wall muscles, causes time-dependent paralysis of wild-type animals. Altered sensitivity to levamisole suggests defects in signaling at the NMJ or muscle function. Here, a protocol for a liquid levamisole assay performed on C. elegans grown in 24-well plates is presented. Vigorous swimming of the animals in liquid allows for the assessment and quantitation of levamisole-induced paralysis in hundreds of worms over a one-hour time period without requiring physical manipulation. This procedure can be used with both wild-type and mutants that have altered sensitivity to levamisole to demonstrate the functional consequences of altered signaling at the NMJ.
Introduction
Activation of postsynaptic acetylcholine receptors (AChRs) on skeletal muscle results in an electrical signal that leads to muscle contraction. Disruption of neuromuscular function results in myasthenic syndromes and muscular dystrophies in humans1,2,3,4. The nematode Caenorhabditis elegans has been used extensively to learn about evolutionarily conserved fundamental biological processes and mechanisms of disease. Vertebrate skeletal muscles and C. elegans body wall muscles are functionally equivalent in the control of locomotion5. Here, a simple assay is presented that can be used to compare wild-type C. elegans with mutants that have altered neuromuscular signaling or muscle function.
Excitatory and inhibitory inputs received from cholinergic and GABAergic motor neurons, respectively, cause muscles on one side of the C. elegans body to contract while the muscles on the other side relax, enabling coordinated locomotion6. Levamisole, an anthelmintic agent used to treat parasitic nematode infections, binds to and constitutively activates one class of AChRs on the body wall muscles, resulting in time-dependent paralysis7. Thus, altered sensitivity to levamisole can be used to identify C. elegans with defects in the balance of excitatory and inhibitory signaling7,8,9,10,11,12,13,14,15. For example, mutations in subunits of the levamisole-sensitive AChR (L-AChR), such as UNC-63, as well as the C. elegans homolog of Cubilin, LEV-10, which is required for L-AChR clustering at the synapse, impact excitatory signaling and result in levamisole resistance7,8. Mutations in the GABAA receptor UNC-49 reduce inhibitory signaling and cause hypersensitivity to levamisole12.
Hypersensitivity or resistance to levamisole has been traditionally assessed by transferring animals to agar plates containing levamisole and then regularly prodding the worms to determine the time point at which paralysis occurs13,14,15. We have developed a liquid levamisole swim assay that eliminates the need for physical manipulation of the animals and allows for the screening of hundreds of animals in just 1 h. Here, the use of this assay with wild-type, unc-63(x26), lev-10(x17), and unc-49(e407) animals is described. However, this protocol can also be performed on C. elegans exposed to RNAi, as was done to validate knockdowns identified in a genome-wide RNAi screen for altered levamisole sensitivity11.
For this pharmacological assay, worms were grown to adulthood in 24-well plates, 0.4 mM levamisole in M9 buffer was added to each well, and the number of moving animals was recorded every 5 min for 1 h. Levamisole-induced paralysis was visually observed over time, and after the completion of the assay, data were quantified. The evaluation of mutants along with wild-type C. elegans allows students to first make predictions about potential phenotypic effects and then perform experiments to test their hypotheses. In conclusion, this simple, inexpensive, liquid levamisole assay is an ideal way to demonstrate the impact of the loss of specific genes on NMJ function.
Protocol
1. Preparation of plates for the levamisole assay
- Prepare nematode growth medium (NGM) media by combining 3 g of NaCl, 2.5 g of peptone, and 17 g of agar with 1 L of deionized (DI) water in a flask with a stir bar.
- After autoclaving, put the media on a hot plate set to 70 °C and stir at a moderate speed for 1 h.
- Add 1 mL of 5 mg/mL cholesterol dropwise to prevent precipitation, 1 mL of 1 M CaCl2, 1 mL of 1 M MgSO4, and 25 mL of 1 M pH 6.0 potassium phosphate buffer to the media.
- Transfer 2 mL of NGM media into each well of a 24-well plate with a sterile serological pipette (Figure 1).
- Allow the plates to dry on the benchtop for 2 days before seeding with bacteria. For longer-term storage, place them at 4 °C.
- Streak out OP50 E. coli on a lysogeny broth (LB) plate and grow overnight at 37 °C.
- Pick a single OP50 colony into B-broth (10 g of tryptone and 5 g of NaCl in 1 L of DI H2O) and set the culture shaking at 37 °C overnight.
- Using a sterile pipet, drop 30 µL of OP50 suspension onto the agar in the middle of each well (Figure 1).
NOTE: Be sure not to damage the agar surface as this will lead to worm burrowing. - Let the plates sit at room temperature for at least 2 days after seeding to allow a bacterial lawn to form.
NOTE: If the bacteria in the center wells is not dry after 2 days, leave the plates open in the hood for 20 min (Figure 1). The plates should be poured and seeded with bacteria 1-2 weeks before the assay.
2. Synchronizing C. elegans (day 1)
- Grow wild-type, unc-63(x26), lev-10(x17), and unc-49(e407) C. elegans to adulthood on 6 cm plates, preparing at least eight plates per strain. Confirm that there are many non-starved gravid adults on the plates.
NOTE: This is twice as many plates as needed; however, it is important to have backup plates in case the first batch of eggs is destroyed during bleaching. - Make 10x M9 buffer by dissolving 59.6 g of Na2HPO4 (dibasic), 29.9 g of KH2PO4 (monobasic), 12.8 g of NaCl, and 2.5 g of MgSO4 in 750 mL of DI water with stirring. Once dissolved, bring the volume up to 1 L and filter sterilize. Dilute 1:10 and autoclave to make 1x M9 buffer.
NOTE: The 1x M9 buffer can be made weeks or months in advance. - Prepare bleaching solution under the hood on the day of synchronization. Mix 10 mL of bleach (Table of Materials), 2.5 mL of 10 N NaOH, and 37.5 mL of DI water in a 50 mL conical tube. Wear goggles, gloves, and a lab coat whenever working with the bleaching solution.
- Using a plastic transfer pipet, wash gravid adult worms from at least four plates with 1x M9 buffer and transfer them into a 15 mL conical tube.
- Spin at 716 x g for 1 min at room temperature, and then remove the supernatant using a transfer pipet.
- Add 10 mL of the bleaching solution. Invert or gently shake the tube for ~4 min until most, but not all, of the worm carcasses have dissolved (Figure 1).
NOTE: Be careful not to over-bleach the worms as this will destroy the eggs. Certain mutations may increase the difficulty of egg isolation. - Spin at 716 x g for 1 min.
- Pour off the bleach solution in one motion; as long as the tube is not shaken at this point, the eggs will stick to the side of the tube.
- Add 15 mL of 1x M9 buffer and invert. Spin at 716 x g for 1 min, and pour off M9 buffer in one smooth motion.
- Perform three washes in total with 1x M9 buffer, as described in step 2.9.
- After the final wash, add 10 mL of fresh 1x M9 and place on a rotator overnight at 15 °C to isolate a synchronized population of starved first larval (L1) stage animals.
NOTE: Before placing on the rotator, check to make sure that there are eggs in the M9 buffer. If not, repeat the prep with backup plates and bleach for a shorter time.
3. Plating synchronized C. elegans (day 2)
- Print out a 24-well plate map and assign strains to randomized places. Each strain should be represented in the 24-well plate at least six times.
- Approximately 24 h after the bleach prep, spin down hatched starved L1 worms in M9 buffer at 716 x g for 1 min at room temperature.
- Remove ~9 mL of M9 buffer with a plastic transfer pipet, and then gently mix the starved first larval stage worms (L1s) in the remaining M9 buffer.
- Immediately pipet 3 µL of the worms in M9 buffer onto a microscope slide and determine the number of L1s; the desired number is 20-30 L1s in 3 µL. Spin down and remove M9 if the worm concentration is too low, and add M9 if the concentration is too high.
NOTE: Check the number of worms per 3 µL at least 2x, inverting the tube each time. - Pipet 3 µL of L1s (20-30 worms total) into each well according to the pre-made plate map (step 3.1; Figure 1).
NOTE: Invert the tube with the L1s in M9 frequently to maintain an even distribution of worms as they will settle to the bottom. If this is not done, some wells will have too few worms, while others will have too many. Also, do not pierce the agar with the pipet tip as this will cause the animals to burrow. - Let the worms grow to adulthood for 3 days at 20 °C.
NOTE: The use of different growth temperatures and certain mutations can impact maturation speed, so be sure to consider the C. elegans life cycle.
4. Performing the levamisole assay (day 5)
- Print a blank datasheet, which will be used to record the number of worms moving in each well every 5 min for 1 h.
NOTE: Students will be able to count the worms in at most 12 wells every 5 min. The top 12 wells are assayed in the first hour, with the bottom 12 wells assayed in the subsequent hour. - Check the worms in the 24-well plates. Using a marker, make an "X" on the plate lid over any wells that have contamination, have starved, or have too many worms (>40), which will make counting difficult.
- Make 0.4 mM levamisole solution by adding 200 µL of 100 mM levamisole stock to 50 mL of 1x M9. Prepare 100 mM levamisole stock by dissolving 240.76 mg of levamisole hydrochloride in 10 mL of H2O and store it at −20 °C.
NOTE: Levamisole must be diluted in M9; dilution in H2O accelerates paralysis. Students should wear gloves. Ingestion of high concentrations of levamisole is toxic. - Start a timer and then, using a transfer pipet, add 1 mL of 0.4 mM levamisole to the first two wells, such that the animals are freely swimming. Continue to add levamisole to the adjacent wells, staggering the time according to the number of wells to be assayed.
NOTE: For example, if 10 wells are assayed, levamisole will be added in the following manner: wells 1 and 2 at time 0, wells 3 and 4 after 1 min, wells 5 and 6 after 2 min, wells 7 and 8 after 3 min, and wells 9 and 10 after 4 min. - At 5 min, start manually counting only the number of moving worms in each well, beginning with the first well, and record that number on the datasheet (Figure 1). Counters can be used but are not required to accurately determine the number of moving worms.
NOTE: Students need to keep an eye on the timer as they will sometimes get behind during early time points before many worms paralyze. The number of time points can be adjusted, or fewer wells can be assayed at each time point if necessary. - Continue to count the number of moving worms in each well every 5 min for 1 h.
NOTE: Immersion in M9 induces constant swimming movement over the course of this 1 h assay, which eliminates the need for the prodding of the worms to assess paralysis (Figure 2A). Exposure to 0.4 mM levamisole solution induces time-dependent paralysis as observed by cessation of swimming; worms that paralyze do not recover. - Repeat steps 4.4.-4.6. for the rest of the wells in the plate.
- At the end of the assay, or when time permits, record the total number of worms in each well.
5. Data analysis
- Obtain the plate map (step 3.1.) and record which strain corresponds to each well.
NOTE: Due to the amount of data collected, the subsequent analysis and discussion of the data will take at least a couple of hours. - Enter the data into a spreadsheet, starting with the total number of worms in each well and organizing by genotype.
- Combining data from the wells, determine the number of worms moving at each time point for every strain.
- Use these data to create a "survival curve" in the statistical software (Table of Materials) to visually display the time-dependent paralysis of the population (Figure 2B).
- Make a data table such that the left-hand column indicates time and subsequent columns contain the data for each strain. For each animal that is paralyzed within the first 5 min, create a row and enter a "1" for 5 min. Repeat this for each time point. For all animals that do not paralyze by the end of the assay, enter a "0" for 60 min.
- Perform pairwise comparisons using the log-rank (Mantel-Cox) test in the statistical software (Table of Materials) to determine if there is a statistically significant difference between two different strains.
- For additional/alternative analysis, instead of performing step 5.3. and step 5.4., determine the percentage of animals moving in each well at every time point. Plot the average with standard error at each time point for all strains assayed using a spreadsheet program or the statistical software (Figure 2C).
Results
Signaling through AChRs and GABA receptors allows muscles on one side of the C. elegans body to contract while the muscles on the other side relax, enabling coordinated locomotion6,16. Levamisole binding to ionotropic L-AChRs causes contraction, leading to time-dependent paralysis of wild-type animals. Here, we assessed the sensitivity of wild type, unc-63 L-AChR mutant, lev-10 L-AChR clustering mutant, and unc-49 GABAA receptor mutant C. elegans to levamisole. Mutations in subunits of the L-AChR, as well as in genes required for trafficking L-AChRs to the muscle plasma membrane, clustering of postsynaptic L-AChRs, and downstream Ca2+ signaling, cause resistance to levamisole-induced paralysis7,8,9,10,11,16,17, as observed in the unc-63 and lev-10 mutants (Figure 2B,C). Loss of the GABA-gated ion channel UNC-49 caused levamisole hypersensitivity (Figure 2B,C) due to disruption of the proper balance of cholinergic and GABAergic signaling12. When this assay was recently performed by undergraduates in an advanced genetics laboratory, 100% of students observed levamisole resistance with the unc-63 and lev-10 mutants, while 88% observed levamisole hypersensitivity with the unc-49 mutant. These data collected with the liquid levamisole swim assay are consistent with phenotypes observed in traditional plate-based levamisole assays7,8,11,12,13.
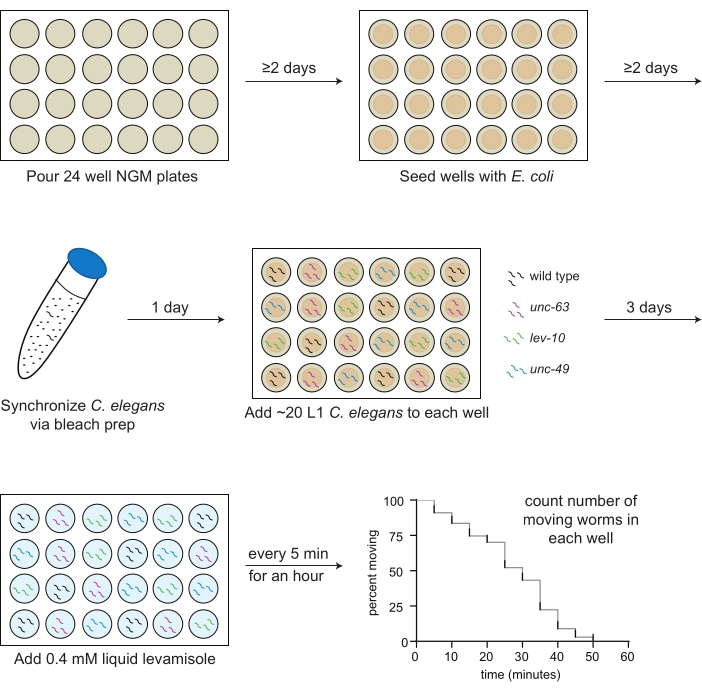
Figure 1: Schematic of levamisole assay preparation and implementation. 24-well NGM plates are poured; 2 days later, OP50 Escherichia coli is seeded into the wells. C. elegans are synchronized through bleach prep, and first larval stage (L1) animals for each strain to be assayed (wild type, black; unc-63(x26), magenta; lev-10(x17), green; unc-49(e407), blue) are pipetted into the wells according to a predetermined plate map the following day. After 3 days of growth at 20 ºC, or when the animals reach adulthood, 0.4 mM levamisole in M9 buffer is added to each well, and the number of moving animals is counted every 5 min for 1 h. Please click here to view a larger version of this figure.
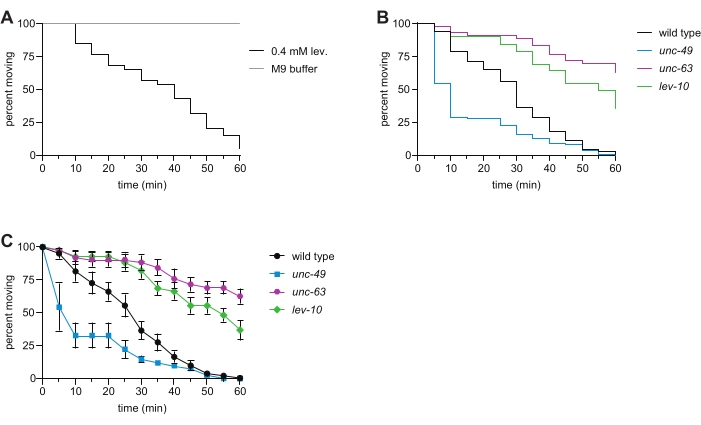
Figure 2: Levamisole-induced paralysis in wild type and representative mutant C. elegans. (A) Exposure to 0.4 mM levamisole causes time-dependent paralysis; this is not observed for animals swimming in 1x M9 buffer for 1 h. (B) Time-dependent paralysis of wild-type (black), unc-63(x26) (magenta), lev-10(x17) (green), and unc-49(e407) (blue) animals in the levamisole swim assay. Loss of the L-AChR subunit UNC-63 or L-AChR clustering protein LEV-10 resulted in levamisole resistance, and loss of the GABAA receptor UNC-49 caused levamisole hypersensitivity. n ≥ 50 per genotype, p < 0.0001 for all mutants compared to the wild type in this representative experiment. (C) Using the same data as in panel (B), the percentage of moving animals at each time point (average of the percentages for each well ± SE) was determined for each strain. Please click here to view a larger version of this figure.
Discussion
Altered response to levamisole can identify genes required for postsynaptic cholinergic signaling and muscle function. The protocol here describes a liquid levamisole assay used to quantitate time-dependent paralysis over a 1 h time period. Students make predictions about the effects of certain genetic mutations, carry out an experiment with large sample size, and then quantitate their results. This assay is a simple, efficient way to quantitate levamisole-induced paralysis without picking or prodding animals, making it suitable for undergraduate laboratories, as well as researchers studying neuromuscular transmission. Discussed here are some of the most common pitfalls, the limitations of the assay, and a variation to the protocol that enables its use with RNAi knockdown animals; also discussed here is how this assay can be embedded into a course-based undergraduate research experience.
Proper preparation of the 24-well plates is essential. First, bacterial lawns must be completely dry before spotting L1 animals in the wells. If not dried enough, the bacteria mixes with the levamisole solution during the assay, turning the liquid cloudy and preventing individuals from being able to count the worms. Second, when synchronizing worms through bleach prep, the animals must not be exposed to the bleach solution for too long as this will cause the protective coating on the eggs to disintegrate. Third, it is crucial to spot only 20-30 L1s per well, as too many worms in the wells make it extremely difficult to accurately count the number moving in the time allotted. Finally, it is important to maintain an aseptic technique during the preparation of the plates as contaminated wells cannot be assayed.
There are a few limitations that must be considered when performing this assay. First, counting the number of moving worms in each well every 5 min may be overwhelming for individuals that lack prior lab experience, and this could lead to imprecise data collection. As for other assays used to determine drug sensitivity18,19, this experiment can be performed with fewer time points. Second, the plating of starved L1s isolated from a bleach prep is an easy method to obtain a synchronized population; however, adjustments must be made if the developmental timing differs between strains being assayed. It is possible to pick late fourth larval stage animals (L4s) into a 24-well plate for this assay; however, when preparing many plates, this is too labor-intensive. Alternatively, one should determine the length of time it takes for each strain to reach L4 and then place the L1s into the wells at different times to adjust for differences in maturation speed. Finally, while this assay can be used to identify new genes important for postsynaptic muscle function11, altered levamisole response can also be caused by mutation or RNAi knockdown of presynaptic genes12. If altered levamisole sensitivity is observed, additional experiments must be performed to determine the reason for this phenotype.
By making a few modifications to the 24-well plates, the described levamisole swim assay can also be performed on RNAi knockdown animals11. Gene knockdown through RNAi can be achieved by feeding C. elegans bacteria that express double-stranded RNA corresponding to the gene of interest20. For the preparation of RNAi plates, 1 mL of 1M IPTG and 1 mL of 25 mg/mL carbenicillin must be added per liter of media after removal from the autoclave. Bacterial cultures are grown by picking a single colony into 3 mL of LB broth plus 3 µL of 25 mg/mLcarbenicillin, and shaking at 37 °C overnight, and the plate map is made when the different bacteria are spotted in the wells. C. elegans are synchronized by bleach prep as described; however, the RNAi hypersensitive eri-1(mg366) strain should be used instead of the wild type to increase gene knockdown. RNAi knockdowns result in the same levamisole phenotypes observed for loss-of-function mutants11.
The protocol presented here can be used in laboratory research, as a stand-alone experiment in the undergraduate lab, or in the opening weeks of an advanced undergraduate course as an introduction to basic C. elegans techniques and neuromuscular function. In a more advanced discovery-based course, students can use this assay to determine if the loss of C. elegans homologs of genes mutated in individuals with myasthenic syndromes, muscular dystrophies, or myopathies alter levamisole sensitivity. In conclusion, this simple, inexpensive levamisole sensitivity assay provides a hands-on approach to learning about signaling at the NMJ and gaining experience working with the C. elegans model system.
Disclosures
The authors have no conflict of interest.
Acknowledgements
We thank Riya Dattani and Lauren Hogstrom, who collected the data in Figure 2B,C blind to genotype in the BISC413 Advanced Genetics Laboratory (Spring 2022) at the University of Delaware. Additional information about the BISC413 course-based undergraduate research experience (CURE), as well as access to the lab manual designed by J. Tanis, is available upon request. Nematode strains were provided by the Caenorhabditis Genetics Center, which is supported by the NIH-ORIP (P40 OD010440). This work was supported by a P20 GM103446 Delaware INBRE Mentored CURE Award (to J.E.T.).
Materials
| Name | Company | Catalog Number | Comments |
| 60 mm non-vented, sharp edge Petri Dishes | TriTech Research | Cat #: T3308 | |
| BD Difco Bacto Agar | Fisher Scientific | Cat#: DF0140-01-0 | |
| BioLite 24 Well Multidish | Fisher Scientific | Cat#:12-556-006 | |
| Calcium Chloride Dihydrate | Fisher Scientific | Cat#: BP510-500 | |
| Cholesterol | MP Biomedicals | Cat#: 02101382-CF | |
| eri-1(mg366) | Caenorhabditis Genetics Center (CGC) | GR1373 | |
| Gibco Bacto Peptone | Fisher Scientific | Cat#: DF0118-17-0 | |
| Gibco Bacto Tryptone | Fisher Scientific | Cat#: DF0123-17-3 | |
| GraphPad Prism (Statistical software) | GraphPad Software, Inc. | ||
| lev-10(x17) | Caenorhabditis Genetics Center (CGC) | ZZ17 | |
| Levamisole hydrochloride | Fisher Scientific | Cat#: AC187870100 | |
| Low Splash Regular Bleach - 121oz - up & up | Target | Search target.com | |
| Magnesium Sulfate Heptahydrate | Fisher Scientific | Cat#: BP213-1 | |
| Microsoft Excel | Microsoft, Inc | ||
| N2 wild type | Caenorhabditis Genetics Center (CGC) | Bristol N2 | |
| OP50 E. coli | Caenorhabditis Genetics Center (CGC) | OP50 | |
| Potassium Phosphate Dibasic | Fisher Scientific | Cat#: BP363-1 | |
| Potassium Phosphate Monobasic | Fisher Scientific | Cat#: BP362-500 | |
| Sodium Chloride (NaCl) | Fisher Scientific | Cat#: BP358-1 | |
| Sodium Hydroxide 10N | Fisher Scientific | Cat#: SS255-1 | |
| Sodium Phosphate Dibasic | Fisher Scientific | Cat#: BP332-1 | |
| unc-49(e407) | Caenorhabditis Genetics Center (CGC) | CB407 | |
| unc-63(x26) | Caenorhabditis Genetics Center (CGC) | ZZ26 |
References
- Engel, A. G., Shen, X. M., Selcen, D., Sine, S. M. Congenital myasthenic syndromes: Pathogenesis, diagnosis, and treatment. The Lancet Neurology. 14 (4), 420-434 (2015).
- Mahjneh, I., Lochmüller, H., Muntoni, F., Abicht, A. DOK7 limb-girdle myasthenic syndrome mimicking congenital muscular dystrophy. Neuromuscular Disorders. 23 (1), 36-42 (2013).
- Rodríguez Cruz, P. M., et al. Congenital myopathies with secondary neuromuscular transmission defects; A case report and review of the literature. Neuromuscular Disorders. 24 (12), 1103-1110 (2014).
- Montagnese, F., et al. Two patients with GMPPB mutation: The overlapping phenotypes of limb-girdle myasthenic syndrome and limb-girdle muscular dystrophy dystroglycanopathy. Muscle and Nerve. 56 (2), 334-340 (2017).
- Gieseler, K., Qadota, H., Benian, G. M. Development, structure, and maintenance of C. elegans body wall muscle. WormBook. , 1-59 (2017).
- Richmond, J. E., Jorgensen, E. M. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nature Neuroscience. 2 (9), 791-797 (1999).
- Lewis, J. A., Wu, C. H., Berg, H., Levine, J. H. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 95 (4), 905-928 (1980).
- Gally, C., Eimer, S., Richmond, J. E., Bessereau, J. L. A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature. 431 (7008), 578-582 (2004).
- Eimer, S., et al. Regulation of nicotinic receptor trafficking by the transmembrane Golgi protein UNC-50. The EMBO Journal. 26 (20), 4313-4323 (2007).
- Gendrel, M., Rapti, G., Richmond, J. E., Bessereau, J. L. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature. 461 (7266), 992-996 (2009).
- Chaya, T., et al. A C. elegans genome-wide RNAi screen for altered levamisole sensitivity identifies genes required for muscle function. G3: Genes, Genomes, Genetics. 11 (4), 047(2021).
- Vashlishan, A. B., et al. An RNAi screen identifies genes that regulate GABA synapses. Neuron. 58 (3), 346-361 (2008).
- Krajacic, P., Pistilli, E. E., Tanis, J. E., Khurana, T. S., Lamitina, S. T. FER-1/Dysferlin promotes cholinergic signaling at the neuromuscular junction in C. elegans and mice. Biology Open. 2 (11), 1245-1252 (2013).
- Gottschalk, A., et al. Identification and characterization of novel nicotinic receptor-associated proteins in Caenorhabditis elegans. The EMBO Journal. 24 (14), 2566-2578 (2005).
- Loria, P. M., Hodgkin, J., Hobert, O. A conserved postsynaptic transmembrane protein affecting neuromuscular signaling in Caenorhabditis elegans. The Journal of Neuroscience. 24 (9), 2191-2201 (2004).
- Treinin, M., Jin, Y. Cholinergic transmission in C. elegans: Functions, diversity, and maturation of ACh-activated ion channels. Journal of Neurochemistry. 158 (6), 1274-1291 (2021).
- Rapti, G., Richmond, J., Bessereau, J. L. A single immunoglobulin-domain protein required for clustering acetylcholine receptors in C. elegans. The EMBO Journal. 30 (4), 706-718 (2011).
- Kim, S., Sieburth, D. A receptor tyrosine kinase network regulates neuromuscular function in response to oxidative stress in Caenorhabditis elegans. Genetics. 211 (4), 1283-1295 (2019).
- Oh, K., Kim, H. Aldicarb-induced paralysis assay to determine defects in synaptic transmission in Caenorhabditis elegans. Bio-Protocol. 7 (14), 2400(2017).
- Timmons, L., Court, D. L., Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263 (1-2), 103-112 (2001).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved