Detection of Bacteriophages in Environmental Samples
Overview
Source: Laboratories of Dr. Ian Pepper and Dr. Charles Gerba - The University of Arizona
Demonstrating Author: Alex Wassimi
Viruses are a unique group of biological entities that infect both eukaryotic and prokaryotic organisms. They are obligate parasites that have no metabolic capacity, and in order to replicate, rely on host metabolism to produce viral parts that self-assemble inside host cells.
Viruses are ultramicroscopic—too small to be viewed with the light microscope, visible only with the greater resolution of the electron microscope. A viral particle consists of a nucleic acid genome, either DNA or RNA, surrounded by a protein coat, known as a capsid, composed of protein subunits or capsomers. In some more complex viruses, the capsid is surrounded by an additional lipid envelope, and some have spike-like surface appendages or tails.
Viruses that infect the intestinal tract of humans and animals are known as enteric viruses. They are excreted in feces and can be isolated from domestic wastewater. Viruses which infect bacteria are known as bacteriophages, and those which infect coliform bacteria are called coliphages (Figure 1). The phages of coliform bacteria are found anywhere coliform bacteria are found.

Figure 1. Coliphage T2.
Principles
Bacteriophages are studied in environmental science because they are a critical component of biological systems. They are the most abundant biological entity on earth and are important because they help control bacterial populations, food web processes, biogeochemical cycles, as well as enhance prokaryotic diversity via horizontal gene transfer. There is also evidence that phages are reliable surrogate indicators for disease-causing enteric viruses that are also fecally-transmitted but difficult to assay. The availability of relatively quick and inexpensive methods to enumerate bacteriophages makes them an attractive tool for the assessment of fecal contamination in environmental samples.
Coliphages in water are assayed by addition of a sample to soft or overlay agar along with a culture of E. coli in the log phase of growth. The phage attach to the bacterial cell and lyse the bacteria. The bacteria produce a confluent lawn of growth except for areas where the phage has grown and lysed the bacteria. These resulting clear areas are known as plaques. A soft agar overlay is used to restrict the physical diffusion of the viruses so that, upon lysing out of a bacterium, they can only spread to neighboring bacterial cells.
To obtain optimal plaque formation it is important that the host bacteria is in the log stage of growth. This ensures that all the phage attach to live bacteria and produce progeny. This requires that a culture of host bacteria be prepared each day that an assay is performed. Usually, a culture is incubated the day before the assay so that it will be in the stationary phase. On the day of the assay, the culture is used to inoculate a broth, which is incubated to obtain enough host bacteria in the log phase for the assay (this usually requires 2-3 h of incubation in a shaking water bath at 35-37 °C).
Procedure
- Obtain a sample of sewage or water containing coliphage.
- Dilute the sample 1:10 and 1:100 using Tris buffer. Do this by transferring 1.0 mL of culture to 9 mL of Tris buffer, and then making a second 10-fold dilution.
- Melt three tubes of soft agar (0.7% nutrient agar or trypticase soy agar per 3 mL tube) by placing them in a steam bath or autoclave.
- Place the agar in a water bath at 45-48 °C for 15 min to allow the temperature of the agar to adjust to 45 °C.
- To the first tube, add 1 mL of a log phase broth culture of E. coli1 and 1 mL of undiluted sample.
- Remove the tube from the water bath and gently rock between hands to mix the suspension for 2-3 s.
- Wipe the water from the tube with a paper towel and pour the agar over a previously prepared Petri dish containing bottom agar (regular nutrient agar or trypticase soy agar).
- Quickly rotate the plate to spread the top agar. Be sure the agar covers the entire surface.
- Repeat Steps 5-8 with the other two tubes of soft agar, using 1 mL of bacteria and 1 mL of each sample dilution (Figure 2).
- After the agar has solidified, invert the Petri dishes and incubate at 37 °C for 48 h. Knock any moisture off the lid of the Petri dish. If a drop of moisture falls on a plaque it will cause the virus to spread across the agar surface.
- After the incubation, count the number of plaques on each dilution (Figure 3) and calculate the concentration of phage in the original sample. Record any major differences in the size or appearance of the plaques.
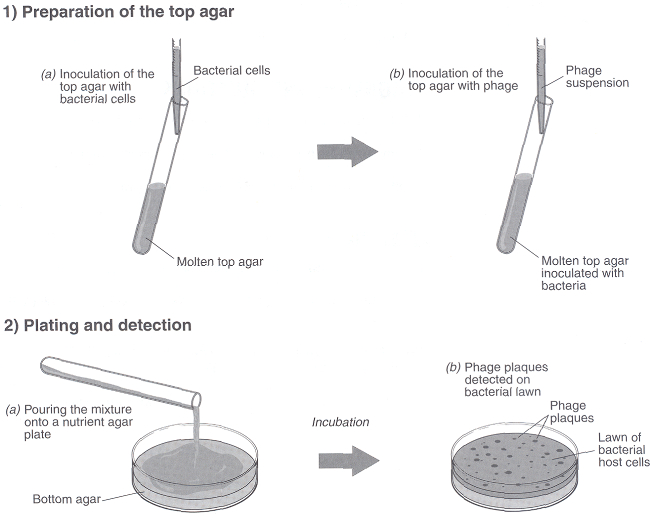
Figure 2. Procedure for the preparation of a bacterial lawn using top agar for coliphage enumeration.
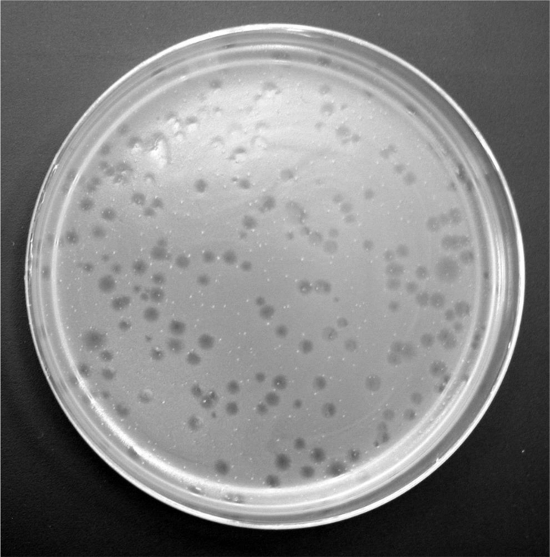
Figure 3. Phage plaques on a bacterial lawn.
1E. coli strain ATCC 15597 usually will produce the greatest number of plaques from sewage samples. It should be grown overnight in a 250-mL Erlenmeyer flask containing 100 mL of nutrient or trypticase soy broth and incubated under shaking conditions at 35 °C. 3 h before the phage assay inoculate one mL of this culture into a fresh flask containing 100 mL of nutrient or trypticase soy broth and place in a shaking water bath at 35-37 °C. This will ensure that the bacteria are in the log phase of growth.
Results
Dilution of sewage sample = 10-1
Number of plaques obtained = 9
Therefore, phage concentration in sewage sample
= 10 x 9 ÷ 1 mL
= 90 plaque-forming units / mL
Raw sewage typically contains 103 – 104 coliphage per mL, with a range of 102 – 108 per mL.
Application and Summary
There are many potential applications of coliphages as environmental indicators. These include their use as indicators of sewage contamination, efficiency of water and wastewater treatment, and survival of enteric viruses and bacteria in the environment. The use of bacteriophages as indicators of the presence and behavior of enteric bacteria and animal viruses has always been attractive because of the ease of detection and low cost associated with phage assays. In addition, they can be quantified in environmental samples within 24 h as compared to days or weeks for enteric viruses.
1E. coli strain ATCC 15597 usually will produce the greatest number of plaques from sewage samples. It should be grown overnight in a 250-mL Erlenmeyer flask containing 100 mL of nutrient or trypticase soy broth and incubated under shaking conditions at 35 °C. 3 h before the phage assay inoculate one mL of this culture into a fresh flask containing 100 mL of nutrient or trypticase soy broth and place in a shaking water bath at 35-37 °C. This will ensure that the bacteria are in the log phase of growth.
Tags
Skip to...
Videos from this collection:

Now Playing
Detection of Bacteriophages in Environmental Samples
Environmental Microbiology
40.6K Views

Determination of Moisture Content in Soil
Environmental Microbiology
358.8K Views

Aseptic Technique in Environmental Science
Environmental Microbiology
126.2K Views

Gram Staining of Bacteria from Environmental Sources
Environmental Microbiology
99.9K Views

Visualizing Soil Microorganisms via the Contact Slide Assay and Microscopy
Environmental Microbiology
42.1K Views

Filamentous Fungi
Environmental Microbiology
57.1K Views

Community DNA Extraction from Bacterial Colonies
Environmental Microbiology
28.8K Views

Detecting Environmental Microorganisms with the Polymerase Chain Reaction and Gel Electrophoresis
Environmental Microbiology
44.5K Views

RNA Analysis of Environmental Samples Using RT-PCR
Environmental Microbiology
40.3K Views

Quantifying Environmental Microorganisms and Viruses Using qPCR
Environmental Microbiology
47.8K Views

Water Quality Analysis via Indicator Organisms
Environmental Microbiology
29.4K Views

Isolation of Fecal Bacteria from Water Samples by Filtration
Environmental Microbiology
39.2K Views

Culturing and Enumerating Bacteria from Soil Samples
Environmental Microbiology
183.7K Views

Bacterial Growth Curve Analysis and its Environmental Applications
Environmental Microbiology
295.5K Views

Algae Enumeration via Culturable Methodology
Environmental Microbiology
13.7K Views
Copyright © 2025 MyJoVE Corporation. All rights reserved