A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
سرعة التحقق من صحة ترمينتورس استخدام الموارد الوراثية النباتية الأزرق البلازميد والجمعية جولدن جيت
In This Article
Summary
This protocol utilizes Golden Gate Assembly and the plasmid pGR-blue to rapidly quantify the strength of terminators found in silico.
Abstract
The goal of this protocol is to allow for the rapid verification of bioinformatically identified terminators. Further, the plasmid (pGR-Blue) is designed specifically for this protocol and allows for the quantification of terminator efficiency. As a proof of concept, six terminators were bioinformatically identified in the mycobacteriophage Bernal13. Once identified, terminators were then made as oligonucleotides with the appropriate sticky ends and annealed together. Using Golden Gate Assembly (GGA), terminators were then cloned into pGR-Blue. Under visible light, false positive colonies appear blue and positively transformed colonies are white/yellow. After induction of an arabinose inducible promoter (pBad) with arabinose, colony strength can be determined by measuring the ratio of green fluorescent protein (GFP) produced to red fluorescent protein (RFP) produced. With pGR-Blue, the protocol can be completed in as little as three days and is ideal in an educational setting. Additionally, results show that this protocol is useful as a means for understanding in silico predictions of terminator efficiency related to the regulation of transcription.
Introduction
Large synthetic biology projects necessitate the use of highly effective transcription terminators to help regulate gene expression. Identification of novel terminators requires bioinformatic analysis of novel genomes. However, as increasing amounts of bioinformatic software are developed, each with a unique algorithm utilized for prediction, more discrepancy between putative results occurs. Because this process is somewhat subjective and is done in silico, these predictions need biological confirmation.1 Additionally, the volume of putative terminators identified through in-silico analysis requires the use of cloning strategies that can be completed in a relatively short time frame.
The PGR-Blue plasmid is a modification of the PGR plasmid that has been redesigned to use Golden Gate Assembly (GGA) to simplify the cloning procedure by allowing for all reaction steps to be simultaneously performed in one micro-centrifuge tube.2,3 Color selection was incorporated into the plasmid to increase the ease of identifying positive colonies. A successful ligation should be white/yellow in visible light and fluoresce green under blue (450 nm) or ultraviolet (UV) light when grown on plates containing arabinose. Because uncut pGR-blue contains a blue chromo protein (amilCP), colonies containing an unmodified plasmid are blue under visible light. This simplification along with the streamlined protocol allows researchers to proceed from bioinformatic identification to biological confirmation in three to four days. The design nature of this system can be beneficial both in the research lab and in educational settings.
The pGR-Blue plasmid allows for quantification of terminator strength.4 A single arabinose inducible promoter is used to produce green fluorescent protein (GFP) and red fluorescent protein (RFP). The terminator is cloned into the plasmid after the GFP sequence but before the RFP sequence, thus stopping the transcription of the RFP protein. The strength of the terminator is determined by the ratio of GFP produced to RFP produced.
The Vision and Change5 report suggested that Science, Technology, Engineering and Math (STEM) education incorporate research based experiences into the classroom.6 However, this requires the development of protocols that can be done by students with limited skill sets in a defined time frame. While the protocol can be accomplished in as little as three days, it was also designed so that each major step could be accomplished in a separate weekly (2-3 hr) lab period to create a Course Research Experience (CRE). When used in this manner, the procedure will take between three and six weeks and is appropriate for both introductory and advanced courses in Genetics, Cell Biology or Bioinformatics.
Protocol
1. تصميم وترتيب أليغنوكليوتيد] مع ينتهي مثبت المناسبة
- تحديد الإنهاء مستقلة رو المحتملة من خلال التحليل الجيني باستخدام البرامج التي هي متاحة بحرية على الانترنت. 7
- عند العمل مع الحمض النووي المزدوج تقطعت بهم السبل، وتحديد اتجاه فاصل لفحصها. 7 البلازميد الموارد الوراثية النباتية-الأزرق يتحقق إلا الإنهاء ligated في 5 'إلى 3' الاتجاه على رأس (حبلا إلى الأمام).
- تحويل حبلا فاصل أسفل (عكس) لتكملة لها عكس لإعادة توجيه تسلسل للاختبار في مكتب المدعي العام الاتحادي الأزرق باستخدام البرمجيات الحرة على الانترنت، على سبيل المثال، القرد.
- بعد تحديد التوجه أن يكون صحيحا، إضافة نهاية لزجة "5'-CGAC-3" إلى 5 'نهاية حبلا العلوي. إضافة نهاية لزجة "5'-CCGC-3" إلى 5 'نهاية حبلا السفلي. 8 وهذا سيضمن سيتم إدراج أن إدراج ligated في المناسبةالتوجه باستخدام GGA.
- مرة واحدة يتم تحديد تسلسل، نظام كما أليغنوكليوتيد] الذين تقطعت بهم السبل واحدة تجاريا.
2. التليين أليغنوكليوتيد] (ينطبق فقط لتجميد المجفف DNA)
- اعادة تعليق [أليغنوكليوتيد الفردية في المياه الحرة نوكلياز إلى تركيز 100 ميكرومتر.
- جعل العازلة 10X الصلب: 1 M كلوريد الصوديوم و 100 ملي تريس، حمض الهيدروكلوريك 7.4 درجة الحموضة. ويمكن تخزين عازلة الصلب في 4 درجات مئوية لعدة أسابيع. مرة واحدة يتم عازلة الصلب وإعادة تعليق أليغنوكليوتيد]، يبدأ الصلب 8.
- لكل فاصل لفحصها، إضافة 16 ميكرولتر من الماء نقي للغاية لأنبوب microcentrifuge 1.5 مل.
- إضافة 2 ميكرولتر من العازلة 10X الصلب إلى كل أنبوب microcentrifuge.
- إضافة 1 ميكرولتر من كبار النوكليوتيد حبلا (100 ميكرومتر) من فاصل المطلوب.
- إضافة 1 ميكرولتر من النوكليوتيد أسفل حبلا (100 ميكرومتر) من فاصل المطلوب.
- إنشاء الغليان با المياهعشر باستخدام الدورق 1000 مل تحتوي على ما يقرب من 400 مل من ماء الصنبور ومكان على طبق ساخن.
- وضع أنابيب microcentrifuge من الخطوة 2.6 في تعويم وتغلى لمدة 4 دقائق. بعد 4 دقائق، إيقاف لوحة الحرارة ولكن ترك الأنبوب وتطفو في حمام مائي لببطء بارد O / N (18 ساعة) 8. تخزين الإنهاء صلب عند درجة حرارة -20 درجة مئوية.
3. الذهبي الجمعية بوابة (ستاندرد ميكس - التكلفة الفعالة)
- جعل 40 نانومتر التخفيف من الإنهاء صلب. إضافة 124 ميكرولتر من المياه nuclease مجانا إلى 1 ميكرولتر من مزيج الإنهاء صلب (100 ميكرومتر).
- عينات الطرد المركزي لمدة 30 ثانية في 10000 x ج ومتجر على الجليد.
- الى ذلك مناسبا أنبوب جديد لthermocycler المستخدمة، إضافة 6 ميكرولتر من المياه nuclease مجانا.
- إضافة 1 ميكرولتر من الحمض النووي التجاري يغاز العازلة [300 ملي تريس، حمض الهيدروكلوريك (الرقم الهيدروجيني 7،7-7،8)، و 100 ملي MgCl 2، 100 ملي DTT و 10 ملي اعبي التنس المحترفين]. تنفيذ جميع الخطوات اللاحقة على الجليد.
- إضافة1 ميكرولتر من الموارد الوراثية النباتية الأزرق (35-50 نانوغرام / ميكرولتر) بلازميد الوجهة إلى أنبوب microcentrifuge.
- إضافة 1 ميكرولتر من 40 نانومتر الإنهاء ملدن إلى أنبوب microcentrifuge.
- إضافة 0.5 ميكرولتر التجاري عالية الدقة BsaI تقييد نوكلياز داخلية للأنبوب microcentrifuge.
- إضافة 0.5 ميكرولتر من يغاز الحمض النووي التجاري إلى أنبوب microcentrifuge.
- أجهزة الطرد المركزي خليط التفاعل في 10000 x ج لمدة 120 ثانية. ثم وضع أنبوب مباشرة في thermocycler.
- تشغيل برنامج thermocycler: 20 دورات من 1 دقيقة عند 37 درجة مئوية تليها 1 دقيقة في 16 درجة مئوية، تليها 1 حلقة 15 دقيقة عند 37 درجة مئوية. استخدام عينات مباشرة أو تجميد وتخزين في درجة حرارة -20 درجة مئوية. 8
4. الذهبي الجمعية بوابة (التجاري ميكس ماجستير - الوقت الفعال)
- إضافة 124 ميكرولتر من المياه nuclease مجانا إلى 1 ميكرولتر من 100 ميكرومتر صلب الإنهاء بإنشائه في الخطوة 2. وهذا يخفف من تركيز الحمض النووي فاصل إلى 40 نانومتر.تنفيذ هذه الخطوة بشكل جديد، وصفت 1.5 مل أنبوب microcentrifuge.
- الى ذلك مناسبا أنبوب جديد لthermocycler، إضافة 14 ميكرولتر من المياه nuclease مجانا. تأكد من وصفت أنبوب جديد وفقا لذلك.
- إضافة 2 ميكرولتر من التجاري ماستر العازلة مزيج إلى أنبوب microcentrifuge.
- إضافة 1 ميكرولتر من الموارد الوراثية النباتية-الأزرق البلازميد (35-50 نانوغرام / ميكرولتر) إلى أنبوب microcentrifuge.
- إضافة 2 ميكرولتر من 40 نانومتر الإنهاء ملدن إلى أنبوب microcentrifuge.
- إضافة 1 ميكرولتر من التجاري ميكس ماجستير في أنبوب microcentrifuge.
- أجهزة الطرد المركزي أنبوب نانومتر 40 في 10000 x ج لمدة 120 ثانية. ثم وضع أنبوب مباشرة في thermocycler.
- تشغيل برنامج thermocycler: 1 حلقة 60 دقيقة عند 37 درجة مئوية تليها 1 حلقة 15 دقيقة في درجة حرارة 55 مئوية. استخدام عينات مباشرة أو تجميد وتخزين في درجة حرارة -20 درجة مئوية.
5. التحول واختيار مستعمرة
- إعداد قاعدة لوريا (LB) أجار لوحات بتري تحتوي على 1 ملم ويركزأيون من الأمبيسلين (الأمبير) وتركيز 10 ملم من أرابينوز (ARB). إعداد لا يقل عن 10 مل من 1 M L حل الأسهم -arabinose لأن هذا الحل الأسهم ستستخدم في خطوات لاحقة.
- استخدام أي عالية الكفاءة (10 أغسطس - 10 سبتمبر transformants / ميكروغرام) المختصة كيميائيا E. القولونية (JM109) خلايا للتحول. 9 للحصول على أفضل النتائج، إضافة 3-5 ميكرولتر من رد فعل ربط إلى 50 ميكرولتر من الخلايا المختصة. عند استخدام الخلايا المختصة الكيميائية، اتبع بروتوكول التحول محددة للخلايا المستخدمة.
- نشر نصف (25 ميكرولتر) من الخلايا تحولت إلى LB قبل تحسنت (أمبير / ARB) أجار لوحات باستخدام على شكل حرف L العقيمة "عصا الهوكي" واحتضان O / N عند 37 درجة مئوية. نظرا لمعدل الأمبيسلين السريع من التحلل، لا احتضان عند 37 درجة مئوية خلال 24 ساعة.
- أداء اختيار مستعمرة على أساس اللون عن طريق التفتيش البصري. وينبغي أن يكون الربط الناجح أبيض / أصفر في الضوء المرئي ويتألق الأخضر undeص الأزرق (450 نانومتر) أو الأشعة فوق البنفسجية. ومن شأن ربط غير ناجحة تنتج مستعمرة هذا هو الازرق في اللون تحت الضوء المرئي بعد 18-20 ساعة.
6. التحقق والكمي لالمنهي
- إعداد 5 مل أنابيب معقمة LB-مرق مع 1 ملم الأمبيسلين و 10 ملي أرابينوز. عدد الأنابيب اللازمة يعتمد على عدد من الإنهاء التي يجري اختبارها بالإضافة إلى ضوابط (خلايا مقاومة الأمبيسلين التي لا يتألق والخلايا التي تحتوي على البلازميد الموارد الوراثية النباتية-الأزرق غير مختصر).
- تحت الضوء المرئي اختيار 2-3 / مستعمرات صفراء البيضاء (الخطوة 5.4) باستخدام حلقة عقيمة. السماح للعينات لاحتضان في المرق على شاكر عند 37 درجة مئوية و 160 دورة في الدقيقة ل16-18 ساعة. لا احتضان هذه العينات لأكثر من 24 ساعة.
- اختياريا، واستخدام مطياف لتحديد كثافة الخلايا الضوئية في 600 نانومتر (OD 600) لضمان نمو الخلايا كاف. عموما، فهناك علاقة OD 600 من ~ 0،8-1،0 بعد 16-18 ساعة من النمو.
- استعمالمعقمة لوحة 96-جيدا لالخطوات المتبقية. إجراء هذه الخطوات في بيئة معقمة وفي ثلاث نسخ. إضافة 199 ميكرولتر من معقم LB + أمبير / الارابينوز مرق (1 ملم الأمبيسلين / 10 ملي الارابينوز) إلى كل بئر. تشمل الغرفة على لوحة لمدة ثلاثة عناصر تحكم منفصلة.
- استخدام الضوابط الثلاثة التالية: بئر (1) التي تحتوي على LB + أمبير / الارابينوز مرق فقط وكذلك (2) للخلايا المختصة هو دون تغيير (فارغ) لتكون بمثابة فارغة. لالكمي (3) من قوة فاصل، ينبغي إدراج الخلايا التي تحتوي على الموارد الوراثية النباتية الأصلية تقطيعه أو التي تحتوي على الموارد الوراثية النباتية الأزرق ligated مع غير إنهاء تسلسل (مراقبة إشارة لتقدير). انظر الشكل رقم 1 لتخطيط لوحة القارئ.
- الماصة 1 ميكرولتر من محلول الخلية من O / N المرق في الآبار الفردية من لوحة 96-جيدا.
- لوحات ختم مع غطاء للتنفس ويهز ل18-20 ساعة عند 37 درجة مئوية و 160 دورة في الدقيقة.
- باستخدام قارئ لوحة، وتحديد OD 600 من خلال قراءةالامتصاصية في 600 نانومتر. قياس GFP مضان كما يلي: الإثارة 395 و 509. الانبعاثات قياس RFP مضان كما يلي: الإثارة 575 والانبعاثات 605. بعد 18-20 ساعة، ينبغي أن OD 600 أن ما يقرب من 0.5.
- لتطبيع للالتباين في النمو، واستخدام المعادلة:
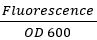 لكلا GFP وطلب تقديم العروض.
لكلا GFP وطلب تقديم العروض. - بعد التطبيع، وتحديد قوة فاصل النسبية باستخدام المعادلة
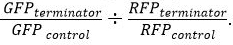
النتائج
وهذا البروتوكول تنتج الخلايا التي تحتوي على الموارد الوراثية النباتية الأزرق مع فاصل ligated بين GFP وطلب تقديم العروض باستخدام الذهبي الجمعية بوابة (الشكل 2). المستعمرات الإيجابية التي تحتوي على إدراج ligated يمكن اختيار على أساس اللون. في الضوء ا...
Discussion
أهم خطوة في هذا البروتوكول هو الصحيح تصميم النوكليوتيد قبل الطلب. يجب أن يكون أليغنوكليوتيد] نهايات لزجة المناسبة إضافة إلى نهايات 5 'من كل من أعلى وأسفل مسارات لضمان GGA التأسيس هو ممكن. بالإضافة إلى ذلك، من المهم أن تبديل اتجاه اليسار الإنهاء التي تواجه (الإنهاء الت...
Disclosures
The authors have nothing to disclose.
Acknowledgements
ان الكتاب أن نعترف مالكوم كامبل وتود Eckdahl مع اتحاد الجينوم لتعليم نشط (GCAT) والتحالف التعليم HHMI-العلوم - الصيادون فج النهوض علم الجينوم وعلم التطوري برنامج (SEA-فاجات).
وقد أيد هذا المشروع من المنح المقدمة من المركز الوطني لبحوث الموارد (P20RR016460) والمعهد الوطني للعلوم الطبية العامة (P20GM103429) من المعاهد الوطنية للصحة. وأيد هذا البحث في جزء من مؤسسة العلوم الوطنية في إطار منحة # IIA-1457888. وقدمت المؤسسية بالإضافة إلى (جامعة أواتشيتا المعمدان) الأموال من خلال دينار باترسون الصيف زمالة أبحاث.
Materials
| Name | Company | Catalog Number | Comments |
| pGR-Blue Plasmid | Addgene | 68374 | |
| pGR-Plasmid | Addgene | 46002 | |
| AeraSeal-(Sterile Sheets) | Excel Scientific | BS-25 | Sterile Sheets only |
| 10X T4 DNA ligase Buffer | NEB | ||
| BsaI-HF | NEB | R3535S | The non-HF enzyme will work but is less heat stable. |
| NEB Golden Gate Assembly Mix | NEB | E1600S | Commerial Master Mix refered to in the protocol. |
| T4 DNA ligase | NEB | M0202S | |
| Round Microcentrifuge Floating Rack | Nova Tech International | F18875-6401 | |
| Ampicillin sodium salt | Sigma Aldrich | A9518 | |
| L-(+)-Arabinose | Sigma Aldrich | A-3256 | D-Arabinose will not induce the pBAD promoter |
| Luria Base (LB) - Broth, Miller | Sigma Aldrich | L1900 | |
| Luria Base (LB) - Agar , Miller | Sigma Aldrich | L2025 | |
| Tecan-Infinite M200 Plate Reader | Tecan | ||
| Mix & Go Competent Cells - Strain JM109 | Zymo Research | T3005 | Use company recommended transformation protocol |
| ApE: A plasmid editor-software | http://biologylabs.utah.edu/jorgensen/wayned/ape/ | ||
| Tris-HCl, Molecular Grade | Promega | H5121 | |
| Sodium Chloride (Crystalline/Biological, Certified) | Fisher Chemical | S671 | |
| Comercial Oligonucleotide synthesis | Integrated DNA Technologies (IDT) | http://www.idtdna.com/site | |
| Microtest Tissue Culture Plates- 96 well (Sterile) | Falcon | 35-3072 | |
| mycobacteriophage "Bernal13" | Genebank | KJ510413 | |
| Nuclease Free Water | Integrated DNA Technologies (IDT) | IDT004 | |
| Sterile, L-shaped Hockey-Stick Cell | Life Science Products | 6444-S1 | |
| Nano-Drop 2000c UV-Vis Spectrometer | Thermo Scientific | 2000c | |
| ARNold: a web tool for the prediction of Rho-independent transcription terminators. | http://rna.igmors.u-psud.fr/ |
References
- Li, J., Zhang, Y. Relationship between promoter sequence and its strength in gene expression. Eur Phys J E Soft Matter. 37 (9), (2014).
- Engler, C., Kandzia, R., Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 3 (11), e3647 (2008).
- Lampropoulos, A., et al. GreenGate---a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One. 8 (12), e83043 (2013).
- Chen, Y. J., et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 10 (7), 659-664 (2013).
- Woodin, T., Carter, V. C., Fletcher, L. Vision and change in biology undergraduate education, a call for action--initial responses. CBE Life Sci Educ. 9 (2), 71-73 (2010).
- Vasaly, H. L., Feser, J., Lettrich, M. D., Correa, K., Denniston, K. J. Vision and change in the biology community: snapshots of change. CBE Life Sci Educ. 13 (1), 16-20 (2014).
- Naville, M., Ghuillot-Gaudeffroy, A., Marchais, A., Gautheret, D. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8 (1), 11-13 (2011).
- Campbell, A. M., et al. pClone: Synthetic Biology Tool Makes Promoter Research Accessible to Beginning Biology Students. CBE Life Sci Educ. 13 (2), 285-296 (2014).
- Rhee, J. I., et al. Influence of the medium composition and plasmid combination on the growth of recombinant Escherichia coli JM109 and on the production of the fusion protein EcoRI::SPA. J Biotechnol. 55 (2), 69-83 (1997).
- Dirla, S., Chien, J. Y., Schleif, R. Constitutive mutations in the Escherichia coli AraC protein. J Bacteriol. 191 (8), 2668-2674 (2009).
- Schleif, R. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 16 (12), 559-565 (2000).
- Kosuri, S., et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci U S A. 110 (34), 14024-14029 (2013).
- Sharon, E., et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 30 (6), 521-530 (2012).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved