需要订阅 JoVE 才能查看此. 登录或开始免费试用。
Method Article
终结者使用PGR-蓝质粒和金门议会的快速验证
摘要
This protocol utilizes Golden Gate Assembly and the plasmid pGR-blue to rapidly quantify the strength of terminators found in silico.
摘要
The goal of this protocol is to allow for the rapid verification of bioinformatically identified terminators. Further, the plasmid (pGR-Blue) is designed specifically for this protocol and allows for the quantification of terminator efficiency. As a proof of concept, six terminators were bioinformatically identified in the mycobacteriophage Bernal13. Once identified, terminators were then made as oligonucleotides with the appropriate sticky ends and annealed together. Using Golden Gate Assembly (GGA), terminators were then cloned into pGR-Blue. Under visible light, false positive colonies appear blue and positively transformed colonies are white/yellow. After induction of an arabinose inducible promoter (pBad) with arabinose, colony strength can be determined by measuring the ratio of green fluorescent protein (GFP) produced to red fluorescent protein (RFP) produced. With pGR-Blue, the protocol can be completed in as little as three days and is ideal in an educational setting. Additionally, results show that this protocol is useful as a means for understanding in silico predictions of terminator efficiency related to the regulation of transcription.
引言
Large synthetic biology projects necessitate the use of highly effective transcription terminators to help regulate gene expression. Identification of novel terminators requires bioinformatic analysis of novel genomes. However, as increasing amounts of bioinformatic software are developed, each with a unique algorithm utilized for prediction, more discrepancy between putative results occurs. Because this process is somewhat subjective and is done in silico, these predictions need biological confirmation.1 Additionally, the volume of putative terminators identified through in-silico analysis requires the use of cloning strategies that can be completed in a relatively short time frame.
The PGR-Blue plasmid is a modification of the PGR plasmid that has been redesigned to use Golden Gate Assembly (GGA) to simplify the cloning procedure by allowing for all reaction steps to be simultaneously performed in one micro-centrifuge tube.2,3 Color selection was incorporated into the plasmid to increase the ease of identifying positive colonies. A successful ligation should be white/yellow in visible light and fluoresce green under blue (450 nm) or ultraviolet (UV) light when grown on plates containing arabinose. Because uncut pGR-blue contains a blue chromo protein (amilCP), colonies containing an unmodified plasmid are blue under visible light. This simplification along with the streamlined protocol allows researchers to proceed from bioinformatic identification to biological confirmation in three to four days. The design nature of this system can be beneficial both in the research lab and in educational settings.
The pGR-Blue plasmid allows for quantification of terminator strength.4 A single arabinose inducible promoter is used to produce green fluorescent protein (GFP) and red fluorescent protein (RFP). The terminator is cloned into the plasmid after the GFP sequence but before the RFP sequence, thus stopping the transcription of the RFP protein. The strength of the terminator is determined by the ratio of GFP produced to RFP produced.
The Vision and Change5 report suggested that Science, Technology, Engineering and Math (STEM) education incorporate research based experiences into the classroom.6 However, this requires the development of protocols that can be done by students with limited skill sets in a defined time frame. While the protocol can be accomplished in as little as three days, it was also designed so that each major step could be accomplished in a separate weekly (2-3 hr) lab period to create a Course Research Experience (CRE). When used in this manner, the procedure will take between three and six weeks and is appropriate for both introductory and advanced courses in Genetics, Cell Biology or Bioinformatics.
研究方案
1.设计和使用适当的粘性末端的寡核苷酸排序
- 确定通过基因组分析使用的是免费提供的在线课程潜在的Rho-独立的终结。7
- 当用双链DNA的工作,决定终止的取向来进行测试。7 PGR-蓝质粒只验证在5结扎'到3'的顶部(正向链)的方向终止子。
- 转换底部(反向)链终止其反向补充,重新调整顺序使用免费在线软件PGR-蓝测试, 如猿。
- 方向被确定为正确后,添加粘底"5'-CGAC-3"'5'顶链的末端。的粘性末端"5'-CCGC-3'"添加到5'和底链的末端。8这将确保连接的插入件将在适当的插入定向使用GGA。
- 一旦序列测定,顺序单链寡核苷酸市场。
2.退火寡核苷酸(仅适用于冻干DNA)
- 重悬在无核酸酶的水个体寡核苷酸的100μM的浓度。
- 使10×退火缓冲液:1M NaCl的和100mM的Tris-HCl pH 7.4的。退火缓冲液可以储存在4℃几周。一旦退火缓冲液是由和寡核苷酸重新悬浮,开始退火8。
- 对于每个要测试的终止子,添加16微升的超纯水至1.5ml微量离心管中。
- 加入2微升10X退火缓冲液中向每个离心管中。
- 从期望终止子加入1μl顶链寡核苷酸(100μM)。
- 从期望终止子加入1μl底链寡核苷酸(100μM)。
- 创建沸水巴次使用含有在热板上大约400毫升自来水和地点的一个1000毫升烧杯中。
- 将步骤2.6在浮动的离心管,煮4分钟。 4分钟后,关火板,但留下的管漂浮在水浴中缓慢冷却O / N(18小时),8。存放于-20℃退火终结。
3.金门大会(标准混合 - 成本效率)
- 使退火终结的40纳米的稀释。加入124μL核酸自由水至1微升退火终结混合(100微米)。
- 离心样品以10,000 xg离心30秒并储存在冰上。
- 为配合新的管适用于热循环,增加6μL核酸自由水。
- 加入1μl商业DNA连接酶缓冲液[300mM的Tris-盐酸(pH值7.7-7.8),100mM的MgCl 2的,100毫摩尔DTT,和10mM ATP]。执行冰的所有后续步骤。
- 加1微升PGR-蓝(35-50纳克/微升)的目标质粒的离心管。
- 加入1μl的40纳米退火终结到离心管中。
- 加入0.5微升商业高保真BsaI限制性内切酶的离心管中。
- 加入0.5微升的商业DNA连接酶的离心管中。
- 离心在10,000rpm xg离心120秒的反应混合物。然后直接放在管进入热循环。
- 运行热循环程序:在37℃下20个循环1分钟,然后以1分钟,在16℃下,随后1个循环的15分钟,在37℃。使用的样品,立即冻结或并储存在-20°C。8
4.金门大会(商业预混 - 时间效率)
- 加入124μL核酸自由水的1微升100μM退火步骤2中创建这削弱了终止DNA浓度至40nm终结的。在执行新的这一步,标示1.5 ml离心管。
- 进入一个新的适合于热循环管中,加入14微升核酸自由水。确保新管相应标记。
- 加入2微升商业预混缓冲液的离心管中。
- 添加PGR-蓝质粒1μL(35-50纳克/微升)的离心管中。
- 加入2微升的40纳米退火终结到离心管中。
- 加入1μl商业预混到离心管中。
- 离心40纳米管在10000 XG 120秒。然后直接放在管进入热循环。
- 运行热循环程序:在37℃1个循环的60分钟,随后1个循环的15分钟在55℃。使用的样品,立即冻结或并储存在-20°C。
5.转型与殖民地的选择
- 准备卢里亚基地(LB)琼脂含1毫米concentrat培养皿氨苄西林离子(AMP)和阿拉伯糖(ARB)的10毫米的浓度。制备至少10毫升1 M L -arabinose原液因为此储备溶液会在后面的步骤中使用。
- 使用任何高效率(10月8日至十月九日转化子/微克)化学感受态大肠杆菌 (JM109)细胞转化。9为了达到最佳效果,加3-5微升连接反应至50μl感受态细胞。当使用化学感受态细胞,遵循针对正在使用的细胞转化协议。
- 传播一半(25微升)的转化的细胞到预热的LB(安培/ ARB)琼脂使用无菌L形"曲棍球"平板上并在37℃孵育O / N。由于氨苄青霉素的分解的速度迅速,不于37℃经24小时孵育。
- 根据通过目测颜色进行菌落筛选。一个成功的结扎应该是白色/可见光黄色和绿色荧光理解过程- [R蓝(450纳米)或紫外光。一个不成功的连接会产生一个殖民地是在18后在可见光下颜色为蓝色 - 20小时。
6.核查和终结者的量化
- 制备5-毫升无菌LB-肉汤管用1mM氨苄青霉素和10mM阿拉伯糖。需要管的数目是依赖于被测试的终止子加对照(细胞以氨苄青霉素抗性不发荧光和含有未切割PGR-蓝质粒的细胞)的数目。
- 在可见光下使用挑无菌环2-3白色/黄色菌落(步骤5.4)。允许样品在肉汤,在37℃和160转16-18小时孵育振荡器上。不孵育这些样品在超过24小时。
- 任选地,使用分光光度计,以确定在600nm(OD 600)的光的细胞密度,以确保有足够的细胞生长。一般来说,〜0.8-1.0的OD 600后增长16-18小时的观察。
- 使用无菌96孔板的其余步骤。进行在无菌环境中,并以一式三份这些步骤。加入199微升无菌LB +安培/阿拉伯糖肉汤(1毫米氨苄西林/ 10毫米阿拉伯糖),每个井。包括在平板上三个独立控制的房间。
- 使用以下三个控件:只有一口井(1)含有LB +安培/阿拉伯糖肉汤和好(2)未转化的感受态细胞(空白)作为空白。用于定量(3)的终止子强度,含有未切割的原始PGR或含PGR-蓝与非终止序列连接单元应包括(为量化参考控制)。参见图1酶标仪布局。
- 吸管1μl的来自澳细胞溶液/ N肉汤到96孔板的各个孔中。
- 密封板具有透气盖,摇晃在37℃18〜20小时,160转。
- 使用读板器,通过读确定OD 600吸光度在600nm处。测量GFP荧光如下:激发395,发射509测量的RFP荧光如下:激发575,发射605 18-20小时后,将OD 600应为大约0.5。
- 为了归在生长变化,使用的公式:
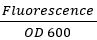 对于GFP和RFP。
对于GFP和RFP。 - 标准化后,确定相对实力终止使用公式
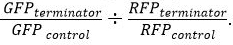
结果
该协议将产生含PGR-蓝用金门组装( 图2)的GFP和RFP之间连接一个终止子的细胞。可以根据颜色来选择含有连接插入阳性菌落。在可见光阳性菌落将白/黄和假阳性将在37℃( 图3)后培养18-20小时产生蓝色菌落。
菌落筛选后,读板器可用于确定终止子的强度。由于概念证明,六公认的终结(T1-6)中的分?...
讨论
在这个协议中最重要的步骤是正确的寡核苷酸设计之前订购。寡核苷酸必须具有适当的粘性末端加入到顶部和底部链二者的5'端,以确保GGA掺入是可能的。此外,为左朝向的终止子(终止子即停止对底链转录)的方向切换到右侧面对它是重要的(终止于顶链转录)终止子,因为GFP和RFP的表达是在正确的朝向(顶链方向)。
两个单独GGA协议和缓冲混合物可以用来在PGR-蓝测试?...
披露声明
The authors have nothing to disclose.
致谢
作者要感谢马尔科姆·坎贝尔和托德Eckdahl与基因组协会的Active教学(GCAT)以及霍华德休斯医学研究所,科学教育联盟 - 噬菌体猎人推进基因组学和进化科学(SEA-噬菌体)计划。
这个项目是由国家研究资源中心(P20RR016460)和通用医学科学院卫生全国学院研究所(P20GM103429)资助。这项研究部分由美国国家科学基金会的资助下#IIA-1457888的支持。此外机构(沃希托浸会大学)资金通过JD帕特森暑期研究奖学金提供。
材料
| Name | Company | Catalog Number | Comments |
| pGR-Blue Plasmid | Addgene | 68374 | |
| pGR-Plasmid | Addgene | 46002 | |
| AeraSeal-(Sterile Sheets) | Excel Scientific | BS-25 | Sterile Sheets only |
| 10X T4 DNA ligase Buffer | NEB | ||
| BsaI-HF | NEB | R3535S | The non-HF enzyme will work but is less heat stable. |
| NEB Golden Gate Assembly Mix | NEB | E1600S | Commerial Master Mix refered to in the protocol. |
| T4 DNA ligase | NEB | M0202S | |
| Round Microcentrifuge Floating Rack | Nova Tech International | F18875-6401 | |
| Ampicillin sodium salt | Sigma Aldrich | A9518 | |
| L-(+)-Arabinose | Sigma Aldrich | A-3256 | D-Arabinose will not induce the pBAD promoter |
| Luria Base (LB) - Broth, Miller | Sigma Aldrich | L1900 | |
| Luria Base (LB) - Agar , Miller | Sigma Aldrich | L2025 | |
| Tecan-Infinite M200 Plate Reader | Tecan | ||
| Mix & Go Competent Cells - Strain JM109 | Zymo Research | T3005 | Use company recommended transformation protocol |
| ApE: A plasmid editor-software | http://biologylabs.utah.edu/jorgensen/wayned/ape/ | ||
| Tris-HCl, Molecular Grade | Promega | H5121 | |
| Sodium Chloride (Crystalline/Biological, Certified) | Fisher Chemical | S671 | |
| Comercial Oligonucleotide synthesis | Integrated DNA Technologies (IDT) | http://www.idtdna.com/site | |
| Microtest Tissue Culture Plates- 96 well (Sterile) | Falcon | 35-3072 | |
| mycobacteriophage "Bernal13" | Genebank | KJ510413 | |
| Nuclease Free Water | Integrated DNA Technologies (IDT) | IDT004 | |
| Sterile, L-shaped Hockey-Stick Cell | Life Science Products | 6444-S1 | |
| Nano-Drop 2000c UV-Vis Spectrometer | Thermo Scientific | 2000c | |
| ARNold: a web tool for the prediction of Rho-independent transcription terminators. | http://rna.igmors.u-psud.fr/ |
参考文献
- Li, J., Zhang, Y. Relationship between promoter sequence and its strength in gene expression. Eur Phys J E Soft Matter. 37 (9), (2014).
- Engler, C., Kandzia, R., Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 3 (11), e3647 (2008).
- Lampropoulos, A., et al. GreenGate---a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One. 8 (12), e83043 (2013).
- Chen, Y. J., et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 10 (7), 659-664 (2013).
- Woodin, T., Carter, V. C., Fletcher, L. Vision and change in biology undergraduate education, a call for action--initial responses. CBE Life Sci Educ. 9 (2), 71-73 (2010).
- Vasaly, H. L., Feser, J., Lettrich, M. D., Correa, K., Denniston, K. J. Vision and change in the biology community: snapshots of change. CBE Life Sci Educ. 13 (1), 16-20 (2014).
- Naville, M., Ghuillot-Gaudeffroy, A., Marchais, A., Gautheret, D. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8 (1), 11-13 (2011).
- Campbell, A. M., et al. pClone: Synthetic Biology Tool Makes Promoter Research Accessible to Beginning Biology Students. CBE Life Sci Educ. 13 (2), 285-296 (2014).
- Rhee, J. I., et al. Influence of the medium composition and plasmid combination on the growth of recombinant Escherichia coli JM109 and on the production of the fusion protein EcoRI::SPA. J Biotechnol. 55 (2), 69-83 (1997).
- Dirla, S., Chien, J. Y., Schleif, R. Constitutive mutations in the Escherichia coli AraC protein. J Bacteriol. 191 (8), 2668-2674 (2009).
- Schleif, R. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 16 (12), 559-565 (2000).
- Kosuri, S., et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci U S A. 110 (34), 14024-14029 (2013).
- Sharon, E., et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 30 (6), 521-530 (2012).
转载和许可
请求许可使用此 JoVE 文章的文本或图形
请求许可探索更多文章
This article has been published
Video Coming Soon
版权所属 © 2025 MyJoVE 公司版权所有,本公司不涉及任何医疗业务和医疗服务。