A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Rapid Verification of Terminators Using the pGR-Blue Plasmid and Golden Gate Assembly
In This Article
Summary
This protocol utilizes Golden Gate Assembly and the plasmid pGR-blue to rapidly quantify the strength of terminators found in silico.
Abstract
The goal of this protocol is to allow for the rapid verification of bioinformatically identified terminators. Further, the plasmid (pGR-Blue) is designed specifically for this protocol and allows for the quantification of terminator efficiency. As a proof of concept, six terminators were bioinformatically identified in the mycobacteriophage Bernal13. Once identified, terminators were then made as oligonucleotides with the appropriate sticky ends and annealed together. Using Golden Gate Assembly (GGA), terminators were then cloned into pGR-Blue. Under visible light, false positive colonies appear blue and positively transformed colonies are white/yellow. After induction of an arabinose inducible promoter (pBad) with arabinose, colony strength can be determined by measuring the ratio of green fluorescent protein (GFP) produced to red fluorescent protein (RFP) produced. With pGR-Blue, the protocol can be completed in as little as three days and is ideal in an educational setting. Additionally, results show that this protocol is useful as a means for understanding in silico predictions of terminator efficiency related to the regulation of transcription.
Introduction
Large synthetic biology projects necessitate the use of highly effective transcription terminators to help regulate gene expression. Identification of novel terminators requires bioinformatic analysis of novel genomes. However, as increasing amounts of bioinformatic software are developed, each with a unique algorithm utilized for prediction, more discrepancy between putative results occurs. Because this process is somewhat subjective and is done in silico, these predictions need biological confirmation.1 Additionally, the volume of putative terminators identified through in-silico analysis requires the use of cloning strategies that can be completed in a relatively short time frame.
The PGR-Blue plasmid is a modification of the PGR plasmid that has been redesigned to use Golden Gate Assembly (GGA) to simplify the cloning procedure by allowing for all reaction steps to be simultaneously performed in one micro-centrifuge tube.2,3 Color selection was incorporated into the plasmid to increase the ease of identifying positive colonies. A successful ligation should be white/yellow in visible light and fluoresce green under blue (450 nm) or ultraviolet (UV) light when grown on plates containing arabinose. Because uncut pGR-blue contains a blue chromo protein (amilCP), colonies containing an unmodified plasmid are blue under visible light. This simplification along with the streamlined protocol allows researchers to proceed from bioinformatic identification to biological confirmation in three to four days. The design nature of this system can be beneficial both in the research lab and in educational settings.
The pGR-Blue plasmid allows for quantification of terminator strength.4 A single arabinose inducible promoter is used to produce green fluorescent protein (GFP) and red fluorescent protein (RFP). The terminator is cloned into the plasmid after the GFP sequence but before the RFP sequence, thus stopping the transcription of the RFP protein. The strength of the terminator is determined by the ratio of GFP produced to RFP produced.
The Vision and Change5 report suggested that Science, Technology, Engineering and Math (STEM) education incorporate research based experiences into the classroom.6 However, this requires the development of protocols that can be done by students with limited skill sets in a defined time frame. While the protocol can be accomplished in as little as three days, it was also designed so that each major step could be accomplished in a separate weekly (2-3 hr) lab period to create a Course Research Experience (CRE). When used in this manner, the procedure will take between three and six weeks and is appropriate for both introductory and advanced courses in Genetics, Cell Biology or Bioinformatics.
Protocol
1. Designing and Ordering Oligonucleotides with the Appropriate Sticky Ends
- Identify potential rho-independent terminators through genomic analysis using programs that are freely available online.7
- When working with double stranded DNA, determine the orientation of the terminator to be tested.7 The pGR-Blue plasmid only verifies terminators ligated in the 5' to 3' direction on the top (forward strand).
- Convert a bottom (reverse) strand terminator to its reversed complement to reorient the sequence for testing in pGR-Blue using free online software, e.g., ApE.
- After the orientation is determined to be correct, add the sticky end " 5'-CGAC-3' " to the 5' end of the top strand. Add the sticky end " 5'-CCGC-3' " to the 5' end of the bottom strand. 8 This will ensure that ligated insert will be inserted in the appropriate orientation using GGA.
- Once sequences are determined, order as single stranded oligonucleotides commercially.
2. Annealing Oligonucleotides (Applies Only to Freeze-dried DNA)
- Re-suspend individual oligonucleotides in nuclease free water to a concentration of 100 µM.
- Make 10x annealing buffer: 1 M NaCl and 100 mM Tris-HCl pH 7.4. Annealing buffer can be stored at 4 °C for several weeks. Once annealing buffer is made and oligonucleotides are re-suspended, begin the annealing8.
- For each terminator to be tested, add 16 µl of ultra-pure water to a 1.5 ml microcentrifuge tube.
- Add 2 µl of 10x annealing buffer to each microcentrifuge tube.
- Add 1 µl of the top strand oligonucleotide (100 µM) from the desired terminator.
- Add 1 µl of the bottom strand oligonucleotide (100 µM) from the desired terminator.
- Create a boiling water bath using a 1,000 ml beaker containing approximately 400 ml of tap water and place on a hot plate.
- Place the microcentrifuge tubes from step 2.6 in a float and boil for 4 min. After 4 min, turn off the heat plate but leave the tube and float in the water bath to slowly cool O/N (18 hr) 8. Store the annealed terminators at -20 °C.
3. Golden Gate Assembly (Standard Mix — Cost Efficient)
- Make a 40 nM dilution of the annealed terminators. Add 124 µl of nuclease free water to 1 µl of annealed terminators mix (100 µM).
- Centrifuge samples for 30 sec at 10,000 x g and store on ice.
- Into a new tube appropriate for the thermocycler being used, add 6 µl of nuclease free water.
- Add 1 µl of commercial DNA Ligase Buffer [300 mM Tris-HCl (pH 7.7-7.8), 100 mM MgCl2, 100 mM DTT, and 10 mM ATP]. Perform all subsequent steps on ice.
- Add 1 µl of pGR-Blue (35-50 ng/µl) destination plasmid to the microcentrifuge tube.
- Add 1 µl of the 40 nM annealed terminators to the microcentrifuge tube.
- Add 0.5 µl commercial high-fidelity BsaI restriction endonuclease to the microcentrifuge tube.
- Add 0.5 µl of commercial DNA ligase to the microcentrifuge tube.
- Centrifuge the reaction mixture at 10,000 x g for 120 sec. Then place the tube directly into the thermocycler.
- Run the thermocycler program: 20 cycles of 1 min at 37 °C followed by 1 min at 16 °C, followed by 1 cycle of 15 min at 37 °C. Use samples immediately or freeze and store at -20 °C.8
4. Golden Gate Assembly (Commercial Master Mix — Time Efficient)
- Add 124 µl of nuclease free water to 1 µl of the 100 µM annealed terminators created in step 2. This dilutes the terminator DNA concentration to 40 nM. Perform this step in a new, labeled 1.5 ml microcentrifuge tube.
- Into a new tube appropriate for the thermocycler, add 14 µl of nuclease free water. Make sure the new tube is labeled accordingly.
- Add 2 µl of Commercial Master Mix Buffer to the microcentrifuge tube.
- Add 1 µl of pGR-Blue plasmid (35-50 ng/µl) to the microcentrifuge tube.
- Add 2 µl of the 40 nM annealed terminators to the microcentrifuge tube.
- Add 1 µl of Commercial Master Mix to the microcentrifuge tube.
- Centrifuge the 40 nM tube at 10,000 x g for 120 sec. Then place the tube directly into the thermocycler.
- Run the thermocycler program: 1 cycle of 60 min at 37 °C followed by 1 cycle of 15 min at 55 °C. Use samples immediately or freeze and store at -20 °C.
5. Transformation and Colony Selection
- Prepare Luria Base (LB) agar Petri plates containing 1 mM concentration of ampicillin (Amp) and a 10 mM concentration of arabinose (arb). Prepare at least 10 ml of 1 M L-arabinose stock solution because this stock solution will be used in later steps.
- Use any high-efficient (108 - 109 transformants/µg ) chemically competent E. coli (JM109) cells for transformation.9 For best results, add 3-5 µl of the ligation reaction to 50 µl of competent cells. When using chemical competent cells, follow the transformation protocol specific to the cells being used.
- Spread half (25 µl) of the transformed cells onto pre-warmed LB (Amp/arb) agar plates using a sterile L-shaped "hockey stick" and incubate O/N at 37 °C. Due to ampicillin's rapid rate of decomposition, do not incubate at 37 °C over 24 hr.
- Perform colony selection based on color by visual inspection. A successful ligation should be white/yellow in visible light and fluoresce green under blue (450 nm) or UV light. An unsuccessful ligation will produce a colony that is blue in color under visible light after 18 - 20 hr.
6. Verification and Quantification of Terminator
- Prepare 5 ml sterile LB-broth tubes with 1 mM ampicillin and 10 mM arabinose. The number of tubes needed is dependent on the number of terminators being tested plus controls (cells with ampicillin resistance that do not fluoresce and cells containing the uncut pGR-Blue plasmid).
- Under visible light pick 2-3 white/yellow colonies (step 5.4) using a sterile loop. Allow the samples to incubate in broths on a shaker at 37 °C and 160 rpm for 16-18 hr. Do not incubate these samples for more than 24 hr.
- Optionally, use a spectrophotometer to determine optical cell density at 600 nm (OD600) to ensure adequate cell growth. Generally, an OD600 of ~0.8-1.0 is observed after 16-18 hr of growth.
- Use a sterile 96-well plate for the remaining steps. Conduct these steps in an aseptic environment and in triplicate. Add 199 µl of sterile LB+ Amp/arabinose broth (1 mM ampicillin/10 mM arabinose) to each well. Include room on the plate for three separate controls.
- Use the following three controls: A well (1) containing LB + Amp/arabinose broth only and a well (2) for untransformed competent cells (blank) to serve as a blank. For quantification (3) of the terminator strength, cells containing uncut original pGR or containing pGR-Blue ligated with a non-terminating sequence should be included (reference control for quantification). See Figure 1 for plate reader layout.
- Pipet 1 µl of the cell solution from the O/N broths into individual wells of the 96-well plate.
- Seal plates with a breathable cover and shake for 18-20 hr at 37 °C and 160 rpm.
- Using a plate reader, determine OD600 by reading absorbance at 600 nm. Measure GFP fluorescence as follows: excitation 395 and emission 509. Measure RFP fluorescence as follows: excitation 575 and emission 605. After 18-20 hr, the OD600 should be approximately 0.5.
- To normalize for variation in growth, use the equation:
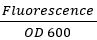 for both GFP and RFP.
for both GFP and RFP. - After normalization, determine relative terminator strength using the equation
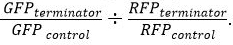
Results
This protocol will produce cells containing pGR-Blue with a terminator ligated between GFP and RFP using Golden Gate Assembly (Figure 2). Positive colonies containing ligated insert can be selected based on color. In visible light positive colonies will be white/yellow and false positives will produce blue colonies after 18-20 hr of incubation at 37 °C (Figure 3).
After colony selection, a ...
Discussion
The most important step in this protocol is proper oligonucleotide design prior to ordering. The oligonucleotides must have the appropriate sticky ends added to the 5' ends of both the top and bottom strands to ensure that GGA incorporation is possible. Additionally, it is important to switch the orientation of left facing terminators (terminators that stop transcription on the bottom strand) to that of right facing (terminates transcription on the top strand) terminators because GFP and RFP expression is in the righ...
Disclosures
The authors have nothing to disclose.
Acknowledgements
Authors would like to acknowledge Malcom Campbell and Todd Eckdahl with the Genome Consortium for Active Teaching (GCAT) and the HHMI-Science Education Alliance - Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program.
This project was supported by grants from the National Center for Research Resources (P20RR016460) and the National Institute of General Medical Sciences (P20GM103429) from the National Institutes of Health. This research was supported in part by the National Science Foundation under grant# IIA-1457888. Additionally institutional (Ouachita Baptist University) funds were provided through the J.D. Patterson Summer Research Fellowship.
Materials
| Name | Company | Catalog Number | Comments |
| pGR-Blue Plasmid | Addgene | 68374 | |
| pGR-Plasmid | Addgene | 46002 | |
| AeraSeal-(Sterile Sheets) | Excel Scientific | BS-25 | Sterile Sheets only |
| 10X T4 DNA ligase Buffer | NEB | ||
| BsaI-HF | NEB | R3535S | The non-HF enzyme will work but is less heat stable. |
| NEB Golden Gate Assembly Mix | NEB | E1600S | Commerial Master Mix refered to in the protocol. |
| T4 DNA ligase | NEB | M0202S | |
| Round Microcentrifuge Floating Rack | Nova Tech International | F18875-6401 | |
| Ampicillin sodium salt | Sigma Aldrich | A9518 | |
| L-(+)-Arabinose | Sigma Aldrich | A-3256 | D-Arabinose will not induce the pBAD promoter |
| Luria Base (LB) - Broth, Miller | Sigma Aldrich | L1900 | |
| Luria Base (LB) - Agar , Miller | Sigma Aldrich | L2025 | |
| Tecan-Infinite M200 Plate Reader | Tecan | ||
| Mix & Go Competent Cells - Strain JM109 | Zymo Research | T3005 | Use company recommended transformation protocol |
| ApE: A plasmid editor-software | http://biologylabs.utah.edu/jorgensen/wayned/ape/ | ||
| Tris-HCl, Molecular Grade | Promega | H5121 | |
| Sodium Chloride (Crystalline/Biological, Certified) | Fisher Chemical | S671 | |
| Comercial Oligonucleotide synthesis | Integrated DNA Technologies (IDT) | http://www.idtdna.com/site | |
| Microtest Tissue Culture Plates- 96 well (Sterile) | Falcon | 35-3072 | |
| mycobacteriophage "Bernal13" | Genebank | KJ510413 | |
| Nuclease Free Water | Integrated DNA Technologies (IDT) | IDT004 | |
| Sterile, L-shaped Hockey-Stick Cell | Life Science Products | 6444-S1 | |
| Nano-Drop 2000c UV-Vis Spectrometer | Thermo Scientific | 2000c | |
| ARNold: a web tool for the prediction of Rho-independent transcription terminators. | http://rna.igmors.u-psud.fr/ |
References
- Li, J., Zhang, Y. Relationship between promoter sequence and its strength in gene expression. Eur Phys J E Soft Matter. 37 (9), (2014).
- Engler, C., Kandzia, R., Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 3 (11), e3647 (2008).
- Lampropoulos, A., et al. GreenGate---a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One. 8 (12), e83043 (2013).
- Chen, Y. J., et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 10 (7), 659-664 (2013).
- Woodin, T., Carter, V. C., Fletcher, L. Vision and change in biology undergraduate education, a call for action--initial responses. CBE Life Sci Educ. 9 (2), 71-73 (2010).
- Vasaly, H. L., Feser, J., Lettrich, M. D., Correa, K., Denniston, K. J. Vision and change in the biology community: snapshots of change. CBE Life Sci Educ. 13 (1), 16-20 (2014).
- Naville, M., Ghuillot-Gaudeffroy, A., Marchais, A., Gautheret, D. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8 (1), 11-13 (2011).
- Campbell, A. M., et al. pClone: Synthetic Biology Tool Makes Promoter Research Accessible to Beginning Biology Students. CBE Life Sci Educ. 13 (2), 285-296 (2014).
- Rhee, J. I., et al. Influence of the medium composition and plasmid combination on the growth of recombinant Escherichia coli JM109 and on the production of the fusion protein EcoRI::SPA. J Biotechnol. 55 (2), 69-83 (1997).
- Dirla, S., Chien, J. Y., Schleif, R. Constitutive mutations in the Escherichia coli AraC protein. J Bacteriol. 191 (8), 2668-2674 (2009).
- Schleif, R. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 16 (12), 559-565 (2000).
- Kosuri, S., et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci U S A. 110 (34), 14024-14029 (2013).
- Sharon, E., et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 30 (6), 521-530 (2012).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved