Coniugazione: un metodo per trasferire la resistenza all'ampicillina dal donatore al ricevente E. coli
Panoramica
Fonte: Alexander S. Gold1, Tonya M. Colpitts1
1 Dipartimento di Microbiologia, Boston University School of Medicine, National Emerging Infections Diseases Laboratories, Boston, MA
Scoperta per la prima volta da Lederberg e Tatum nel 1946, la coniugazione è una forma di trasferimento genico orizzontale tra batteri che si basa sul contatto fisico diretto tra due cellule batteriche (1). A differenza di altre forme di trasferimento genico, come la trasformazione o la trasduzione, la coniugazione è un processo naturale in cui il DNA viene secreto da una cellula donatrice a una cellula ricevente in modo unidirezionale. Questa direzionalità e la capacità di questo processo di aumentare la diversità genetica dei batteri ha dato alla coniugazione la reputazione di una forma di "accoppiamento" batterico, che si ritiene abbia contribuito notevolmente al recente aumento dei batteri resistenti agli antibiotici (2, 3). Utilizzando pressioni selettive, ad esempio l'uso di antibiotici, la coniugazione è stata manipolata per l'uso in laboratorio, rendendola un potente strumento per il trasferimento genico orizzontale tra batteri e, in alcuni casi, da batteri a lieviti, cellule vegetali e animali (4). Oltre alle applicazioni in laboratorio, il trasferimento genico batterio-eucariota mediante coniugazione è un'entusiasmante via di trasferimento del DNA con una moltitudine di possibili applicazioni biotecniche e implicazioni naturali (5).
Si pensa che la coniugazione funzioni con un "meccanismo a due fasi" (6). In primo luogo, prima che qualsiasi DNA possa essere trasferito, la cellula donatrice deve entrare in contatto diretto cellula a cellula con il ricevente. Questo processo è stato caratterizzato meglio nei batteri gram-negativi, il più studiato dei quali è Escherichia coli. Il contatto cellula-cellula è stabilito dalla presenza di una complessa rete di filamenti extracellulari sul donatore nota come sex pilus, un elemento coniugativo codificato dal gene trasferibile noto come fattore F (fertilità) (7, 8). Oltre a stabilire un contatto tra donatore e ricevente, diverse proteine vengono trasportate attraverso il pilus sessuale al citoplasma ricevente, formando un condotto del sistema di secrezione di tipo IV (T4SS) tra le due cellule, una struttura necessaria per la seconda fase di coniugazione, il trasferimento del DNA (6). Combinando questa funzione del pilus sessuale con la replicazione del cerchio rotolante del DNA, la cellula donatrice è in grado di trasferire il DNA sotto forma di un elemento trasponibile, come un plasmide o un trasposone, al ricevente mediante un modello "shoot and pump" (6). In questo caso, lo "shooting" è il trasporto della proteina pilota, con DNA collegato, da parte del T4SS nella cellula ricevente, e il "pumping" è il trasporto attivo del DNA al ricevente, un processo dipendente dal T4SS e catalizzato dall'accoppiamento delle proteine (6). Il macchinario utilizzato in questo processo è composto da un'origine di sequenza di trasferimento (oriT), che deve essere fornita dal DNA nei geni cis e trans, che codificano una relaxasi, un complesso di formazione di coppie di accoppiamenti e una proteina di accoppiamento di tipo IV, e possono essere presenti in cis o trans (9). Questa relaxasi fende il sito nic all'interno della sequenza oriT e si attacca covalentemente all'estremità 5' del filamento trasferito per produrre il relaxosoma, un complesso DNA-relaxasi a singolo filamento con altre proteine ausiliarie (9). Una volta formato, il relaxosoma si collega al complesso di formazione della coppia di accoppiamento, tramite la proteina di accoppiamento di tipo IV, che consente il trasferimento del complesso ssDNA-relaxasi nelle cellule riceventi da parte del T4SS (10). Una volta nel citoplasma del ricevente, il DNA può integrarsi nel genoma del ricevente o esistere separatamente sotto forma di plasmide, entrambi i quali consentono l'espressione dei suoi geni.
In questo esperimento, il ceppo di donatore di coniugazione ampiamente utilizzato E. coli WM3064 è stato utilizzato per trasferire il gene che codifica per la resistenza all'ampicillina al ceppo ricevente E. coli J53. Mentre entrambi i ceppi dei batteri gram-negativi erano resistenti alla tetraciclina, solo il ceppo donatore WM3064 aveva il gene per la resistenza all'ampicillina, codificato per nel vettore navetta pWD2-oriT, ed era auxotrofico all'acido diaminopimetico (DAP) (11-13). Questo esperimento consisteva in due fasi principali, la preparazione di ceppi donatore e ricevente, seguita dal trasferimento del gene di resistenza all'ampicillina dal donatore al ricevente mediante coniugazione (Figura 1).
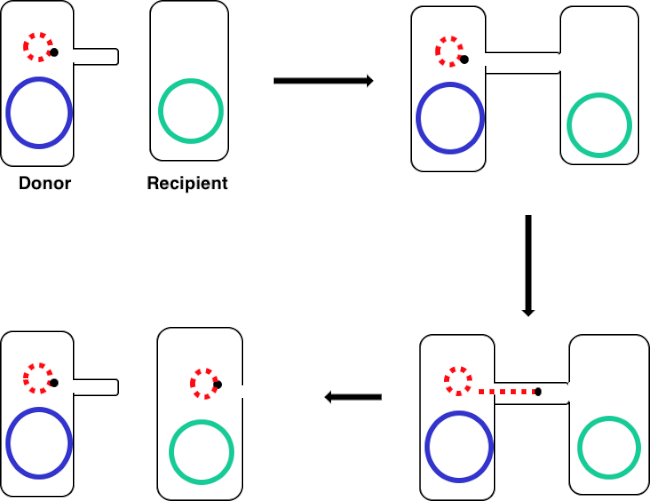
Figura 1: Schema di coniugazione. Questo schema mostra il successo del trasferimento di un plasmide, solo un esempio di elemento di DNA trasponibile, da una cellula donatrice a una cellula ricevente usando la coniugazione. Al contatto con la cellula ricevente da parte della cellula donatrice attraverso il pilus sessuale, il plasmide si replica mediante replicazione del cerchio rotolante, si muove attraverso il complesso multiproteico unendo le due cellule e forma un nuovo plasmide a lunghezza intera nella cellula ricevente.
Incubando una miscela di cellule donatrici e riceventi, quindi placcando successivamente queste cellule in presenza di tetraciclina e DAP, ciò ha permesso il trasferimento riuscito del gene di resistenza all'ampicillina. Successivamente, le cellule placcate cresciute da questa miscela in presenza di tetraciclina e ampicillina, hanno rimosso tutte le cellule donatrici a causa della mancanza di DAP e di eventuali cellule riceventi che potrebbero non aver acquisito il gene di resistenza all'ampicillina, producendo batteri del ceppo J53 strettamente riceventi che hanno acquisito resistenza all'ampicillina (Figura 2). Una volta effettuato, il successo del trasferimento del gene di resistenza all'ampicillina è stato confermato dalla PCR. Poiché la coniugazione ha avuto successo, il ceppo J53 di E. coli conteneva pWD2-oriT ed era resistente all'ampicillina, e il gene che codifica per questa resistenza è rilevabile dalla PCR. Tuttavia, in caso di insuccestamento non ci sarebbe stato alcun rilevamento del gene di resistenza all'ampicillina e l'ampicillina avrebbe comunque funzionato come un antibiotico efficace contro il ceppo J53.
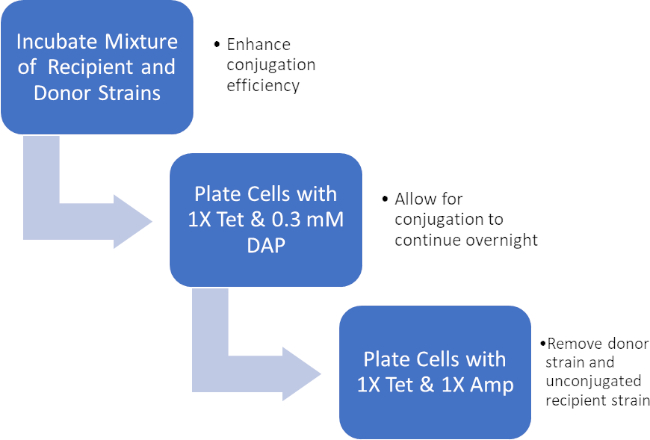
Figura 2: Schema del protocollo. Questo schema mostra una panoramica del protocollo presentato.
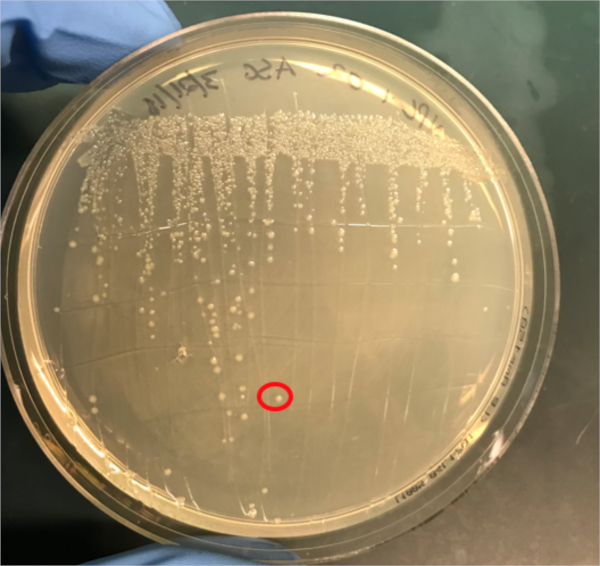
Figura 3A: La conferma del successo della coniugazione mediante PCR. A) Le scorte congelatrici dei campioni di controllo coniugati e negativi sono state strisciate su piastre di agar e una colonia è stata selezionata (rossa) per l'isolamento del DNA.
Procedura
1. Configurazione
- Autoclave circa 1L di Luria-Bertani medium (LB). Questo LB sterile sarà utilizzato per produrre circa 5 ml di LB contenente 0,3 mM di acido diaminopimetico (DAP).
- Raccogliere le seguenti piastre: piastre di agar LB con 1X Tet e 0,3 mM DAP, piastre lb agar con 1X Tet solo e piastre LB agar con solo 1X Amp/Tet.
- Assicurarsi che un po 'di glicerolo e una scatola di punte di pipette di plastica pre-sterilizzate siano a portata di mano.
- Prima di iniziar
Risultati
Se la coniugazione ha avuto successo, un prodotto PCR a banda di 500 coppie di basi sarà osservato nel pozzo in cui è stata caricata la reazione PCR 1 (Ben #2 in Figura 3B), mentre non si osserveranno bande nel pozzo in cui è stata caricata la reazione PCR 3 (Ben #4 in Figura 3B). La presenza di questa banda conferma il successo del trasferimento del gene di resistenza all'ampicillina, conferendo così resistenza all'ampicillina al ceppo J53 di E. coli.
Log in or to access full content. Learn more about your institution’s access to JoVE content here
Applicazione e Riepilogo
La coniugazione è un processo naturale di trasferimento genico orizzontale che si basa sul contatto diretto cellula-cellula di una cellula donatrice e di una cellula ricevente. Questo processo è condiviso tra tutti i tipi di batteri ed è stato determinante nell'evoluzione batterica, in particolare la resistenza agli antibiotici. In laboratorio, la coniugazione può essere utilizzata come metodo efficace di trasferimento genico che è molto meno dirompente rispetto ad altre tecniche. Al di fuori del laboratorio, la cap...
Riferimenti
- Lederberg J, Tatum, E.L. Gene recombination in Escherichia coli Nature. 1946;158:558.
- Holmes R.K. J, M.G. Genetics: Exchange of Genetic Information. 4th Edition ed. Baron S, editor. Galveston, TX: University of Texas Medical Branch at Galveston; 1996.
- Cruz F, Davies, J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends in Microbiology. 2000;8:128-33.
- Llosa M, Cruz, F. Bacterial conjugation: a potential tool for genomic engineering. Ressearch in Microbiology. 2005;156:1-6.
- Lacroix B, Citovsky, V. Transfer of DNA from Bacteria to Eukaryotes. mBio. 2016;7(4):1-9.
- Llosa M, et al. Bacterial conjugation: a two-step mechanism for
- DNA transport. Molecular Microbiology. 2002;45:1-8.
- Grohmann E, Muth, G., Espinosa, M. Conjugative Plasmid Transfer in Gram-Positive Bacteria. Microbiology and Molecular Biology Reviews. 2003;67:277-301.
- Firth N, Ippen-Ihler, K, Skurray, RA. Structure and function of the F factor and mechanism of conjugation. Escherichia coli and salmonella: cellular and molecular biology. 1996;2:2377-401.
- Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EPC, De La Cruz F. Mobility of Plasmids. Microbiology and Molecular Biology Reviews. 2010;74(3):434-52.
- Cascales E. Definition of a Bacterial Type IV Secretion Pathway for a DNA Substrate. 2004;304(5674):1170-3.
- Wang P, Yu Z, Li B, Cai X, Zeng Z, Chen X, et al. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microbial Cell Factories. 2015;14(1):11.
- Yi H, Cho YJ, Yong D, Chun J. Genome Sequence of Escherichia coli J53, a Reference Strain for Genetic Studies. Journal of Bacteriology. 2012;194(14):3742-3.
- Baumann RLB, E. H.; Wiseman, J. S.; Vaal, M.; Nichols, J. S. Inhibition of Escherichia coli Growth and Diaminopimelic Acid Epimerase by 3-Chlorodiaminopimelic Acid. Antimicrobial Agents and Chemotherapy 1988;32:1119-23.
- Rocha D, Santos, CS, Pacheco LG. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685-93.
Vai a...
Video da questa raccolta:

Now Playing
Coniugazione: un metodo per trasferire la resistenza all'ampicillina dal donatore al ricevente E. coli
Microbiology
38.2K Visualizzazioni

Creazione di una colonna di Winogradsky: un metodo per arricchire le specie microbiche presenti in un campione di sedimento
Microbiology
129.4K Visualizzazioni

Diluizioni seriali e piastratura: la conta microbica
Microbiology
316.1K Visualizzazioni

Culture di arricchimento: coltura di microbi aerobici e anaerobici su terreni selettivi e differenziali
Microbiology
132.1K Visualizzazioni

Colture pure e piastratura per striscio: isolamento di singole colonie batteriche da un campione misto
Microbiology
166.2K Visualizzazioni

Sequenziamento dell'rRNA 16S: una tecnica basata sulla PCR per identificare le specie batteriche
Microbiology
189.0K Visualizzazioni

Curve di crescita: generazione di curve di crescita utilizzando le unità formanti colonia e la misurazione della densità ottica
Microbiology
296.0K Visualizzazioni

Test di suscettibilità agli antibiotici: test dell'epsilometro per determinare i valori MIC di due antibiotici e valutare la sinergia antibiotica
Microbiology
93.8K Visualizzazioni

Microscopia e colorazioni: la colorazione di Gram, delle endospore e del capside
Microbiology
363.3K Visualizzazioni

Saggio delle placche: un metodo per determinare il titolo virale in unità formanti placca (UFP)
Microbiology
186.2K Visualizzazioni

Trasformazione di cellule di E. coli tramite l'utilizzo di una procedura basata sul metodo del cloruro di calcio
Microbiology
86.8K Visualizzazioni

La trasduzione batterica tramite fagi: un metodo per trasferire la resistenza all'ampicillina da una cellula donatore di E. coli ad una ricevente
Microbiology
29.1K Visualizzazioni