La trasduzione batterica tramite fagi: un metodo per trasferire la resistenza all'ampicillina da una cellula donatore di E. coli ad una ricevente
Panoramica
Fonte: Alexander S. Gold1, Tonya M. Colpitts1
1 Dipartimento di Microbiologia, Boston University School of Medicine, National Emerging Infections Diseases Laboratories, Boston, MA
La trasduzione è una forma di scambio genetico tra batteri che utilizza batteriofagi, o fagi, una classe di virus che infetta esclusivamente organismi procariotici. Questa forma di trasferimento del DNA, da un batterio all'altro attraverso un fago, fu scoperta nel 1951 da Norton Zinder e Joshua Ledererg, che definirono il processo "trasduzione" (1). I batteriofagi furono scoperti per la prima volta nel 1915 dal batteriologo britannico Frederick Twort, poi scoperti indipendentemente nel 1917 dal microbiologo franco-canadese Felix d'Herelle (2). Da allora, la struttura e la funzione di questi fagi sono state ampiamente caratterizzate (3), dividendo questi fagi in due classi. La prima di queste classi sono i fagi litici che al momento dell'infezione si moltiplicano all'interno del batterio ospite, interrompendo il metabolismo batterico, lisi della cellula e rilasciando fagi di progenie (4). Come risultato di questa attività antibatterica e della crescente prevalenza di batteri resistenti agli antibiotici, questi fagi litici si sono recentemente dimostrati utili come trattamento sostitutivo degli antibiotici. La seconda di queste classi sono i fagi lisogenici che possono moltiplicarsi all'interno dell'ospite attraverso il ciclo litico o entrare in uno stato quiescente in cui il loro genoma è integrato in quello dell'ospite (Figura 1), un processo noto come lisogenesi, con la capacità di indotta dalla produzione di fagi in più generazioni successive (4).
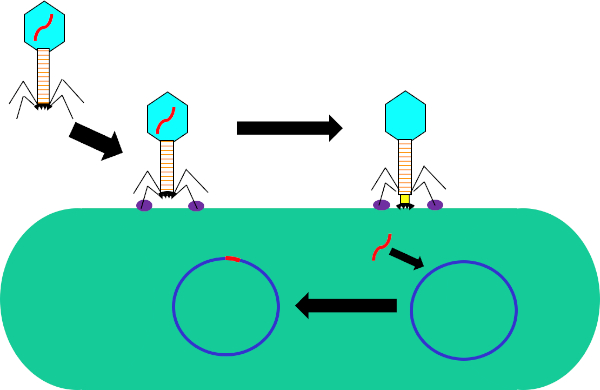
Figura 1: Infezione della cellula ospite da parte del batteriofago. Adsorbimento da parte del fago alla parete cellulare batterica attraverso interazioni tra le fibre della coda e il recettore (viola). Una volta sulla superficie cellulare, il fago viene irreversibilmente attaccato alla cellula batterica usando la piastra di base (nera) che viene spostata sulla parete cellulare dalla guaine contrattile (gialla). Il genoma dei fagi (rosso) entra quindi nella cellula e si integra nel genoma della cellula ospite.
Mentre la trasduzione batterica è un processo naturale, utilizzando la tecnologia moderna è stata manipolata per il trasferimento di geni nei batteri in laboratorio. Inserendo geni di interesse nel genoma di un fago lisogenico, come il fago, si è in grado di trasferire questi geni nei genomi dei batteri e di conseguenza esprimerli all'interno di queste cellule. Mentre altri metodi di trasferimento genico, come la trasformazione, utilizzano un plasmide per il trasferimento e l'espressione genica, l'inserimento del genoma del fago in quello del batterio ricevente non solo ha il potenziale per conferire nuovi tratti a questo batterio, ma consente anche mutazioni naturali e altri fattori dell'ambiente cellulare per alterare la funzione del gene trasferito.
Rispetto ad altri metodi di trasferimento genico orizzontale, come la coniugazione, la trasduzione è abbastanza flessibile nei criteri richiesti per le cellule donatrici e riceventi. Qualsiasi elemento genetico che può adattarsi all'interno del genoma del fago utilizzato può essere trasferito da qualsiasi ceppo di batteri donatori a qualsiasi ceppo di batteri riceventi purché entrambi siano permissivi al fago, richiedendo l'espressione dei necessari recettori fagici sulle superfici cellulari. Una volta che questo gene viene spostato fuori dal genoma del donatore e confezionato nel fago, può essere trasferito al ricevente. Dopo la trasduzione, è necessario selezionare per i batteri riceventi che contengono il gene di interesse devono essere selezionati per. Questo potrebbe essere fatto utilizzando un marcatore genetico, come un FLAG-tag o un polyhistidine-tag, per contrassegnare il gene di interesse, o la funzione intrinseca del gene, nel caso di geni che codificano per la resistenza agli antibiotici. Inoltre, la PCR potrebbe essere utilizzata per confermare ulteriormente il successo della trasduzione. Utilizzando primer per una regione all'interno del gene di interesse e confrontando il segnale con un controllo positivo, batteri che hanno il gene di interesse e un controllo negativo, batteri che hanno subito gli stessi passaggi della reazione di trasduzione senza fagi. Mentre la trasduzione batterica è uno strumento utile in biologia molecolare, ha e continua a svolgere un ruolo importante nell'evoluzione dei batteri, in particolare per quanto riguarda il recente aumento della resistenza agli antibiotici.
In questo esperimento, la trasduzione batterica è stata utilizzata per trasferire il gene che codifica per la resistenza all'ampicillina antibiotica dal ceppo W3110 di E.coli al ceppo J53 attraverso il batteriofago P1 (5). Questo esperimento consisteva in due fasi principali. In primo luogo, la preparazione del fago P1 contenente il gene di resistenza all'ampicillina dal ceppo donatore. In secondo luogo, il trasferimento di questo gene al ceppo ricevente per trasduzione con il fago P1 (Figura 1). Una volta effettuato, il successo del trasferimento del gene di resistenza all'ampicillina potrebbe essere determinato dalla qPCR (Figura 2). Se la trasduzione ha successo, il ceppo J53 di E. coli sarebbe resistente all'ampicillina e il gene che conferisce questa resistenza rilevabile dalla qPCR. In caso di insuccesta, non ci sarebbe alcun rilevamento del gene di resistenza all'ampicillina e l'ampicillina funzionerebbe ancora come un antibiotico efficace contro il ceppo J53.
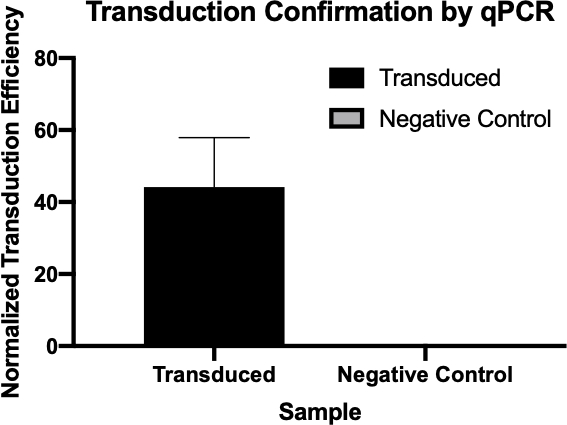
Figura 2: La conferma del successo della trasduzione da parte della qPCR. Confrontando i valori di Cq rilevati per il gene di interesse dalla reazione di trasduzione e dalla reazione di controllo negativa e normalizzando questi valori con un gene di pulizia, si è stati in grado di confermare che la trasduzione batterica ha avuto successo.
Procedura
1. Configurazione
- Prima di iniziare qualsiasi lavoro che coinvolga i microbi, sterilizzare lo spazio di lavoro utilizzando il 70% di etanolo. Utilizzare sempre i DPI necessari (camice da laboratorio e guanti).
- Assicurarsi che le piastre di agar LB con 1x ampicillina, soluzione di lisato fagico P1 disponibile in commercio, cloroformio, citrato di sodio 1 M, glicerolo e una scatola di punte di pipette di plastica pre-sterilizzate e diffusori di cellule siano a portata di mano.
Applicazione e Riepilogo
Il trasferimento di geni da e verso i batteri da parte del batteriofago, pur essendo un processo naturale, si è rivelato estremamente utile per una moltitudine di scopi di ricerca. Mentre sono possibili altri metodi di trasferimento genico come la trasformazione e la coniugazione, la trasduzione utilizza in modo univoco i batteriofagi; non solo consentendo l'integrazione genica nel genoma dell'ospite, ma anche la consegna di geni a più batteri che non sono suscettibili ad altri metodi. Questo processo, sebbene particol..
Riferimenti
- Lederberg J, Lederberg E.M., Zinder, N.D., et al. Recombination analysis of bacterial heredity. Cold Spring Harbor symposia Quantitative Biol. 1951;16:413-43.
- Duckworth DH. "Who Discovered Bacteriophage?". Bacteriology Reviews. 1976;40:793-802.
- Yap ML, Rossman, M.G. Structure and Function of Bacteriophage T4. Future Microbiol. 2014;9:1319-27.
- Sulakvelidze A, Alavidze, Z., Morris, J. G. Bacteriophage Therapy Antimicrobial Agents and Chemotherapy 2001;45(3):649-59.
- Moore S. Sauer:P1vir phage transduction 2010 [Available from: https://openwetware.org/wiki/Sauer:P1vir_phage_transduction].
- Kobayashi A, et al. Growth Phase-Dependent Expression of Drug Exporters in
- Escherichia coli and Its Contribution to Drug Tolerance. Journal of Bacteriology. 2006;188(16):5693-703.
- Rocha D, Santos, CS, Pacheco LG. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685-93.
- Pálffy R. et al. Bacteria in gene therapy: bactofection versus alternative gene therapy. Gene Ther. 2006 13:101-5.
- O'Neill JM, et al. Intestinal delivery of non-viral gene therapeutics: physiological barriers and preclinical models. Drug Discovery Today. 2011;16:203-2018.
Vai a...
Video da questa raccolta:

Now Playing
La trasduzione batterica tramite fagi: un metodo per trasferire la resistenza all'ampicillina da una cellula donatore di E. coli ad una ricevente
Microbiology
29.1K Visualizzazioni

Creazione di una colonna di Winogradsky: un metodo per arricchire le specie microbiche presenti in un campione di sedimento
Microbiology
129.9K Visualizzazioni

Diluizioni seriali e piastratura: la conta microbica
Microbiology
317.0K Visualizzazioni

Culture di arricchimento: coltura di microbi aerobici e anaerobici su terreni selettivi e differenziali
Microbiology
132.2K Visualizzazioni

Colture pure e piastratura per striscio: isolamento di singole colonie batteriche da un campione misto
Microbiology
166.4K Visualizzazioni

Sequenziamento dell'rRNA 16S: una tecnica basata sulla PCR per identificare le specie batteriche
Microbiology
189.6K Visualizzazioni

Curve di crescita: generazione di curve di crescita utilizzando le unità formanti colonia e la misurazione della densità ottica
Microbiology
297.8K Visualizzazioni

Test di suscettibilità agli antibiotici: test dell'epsilometro per determinare i valori MIC di due antibiotici e valutare la sinergia antibiotica
Microbiology
93.9K Visualizzazioni

Microscopia e colorazioni: la colorazione di Gram, delle endospore e del capside
Microbiology
364.0K Visualizzazioni

Saggio delle placche: un metodo per determinare il titolo virale in unità formanti placca (UFP)
Microbiology
186.5K Visualizzazioni

Trasformazione di cellule di E. coli tramite l'utilizzo di una procedura basata sul metodo del cloruro di calcio
Microbiology
87.1K Visualizzazioni

Coniugazione: un metodo per trasferire la resistenza all'ampicillina dal donatore al ricevente E. coli
Microbiology
38.4K Visualizzazioni