Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Thermodynamics of Membrane Protein Folding Measured by Fluorescence Spectroscopy
W tym Artykule
Podsumowanie
This video article details the experimental procedure for obtaining the Gibbs free energy of membrane protein folding by tryptophan fluorescence.
Streszczenie
Membrane protein folding is an emerging topic with both fundamental and health-related significance. The abundance of membrane proteins in cells underlies the need for comprehensive study of the folding of this ubiquitous family of proteins. Additionally, advances in our ability to characterize diseases associated with misfolded proteins have motivated significant experimental and theoretical efforts in the field of protein folding. Rapid progress in this important field is unfortunately hindered by the inherent challenges associated with membrane proteins and the complexity of the folding mechanism. Here, we outline an experimental procedure for measuring the thermodynamic property of the Gibbs free energy of unfolding in the absence of denaturant, ΔG°H2O, for a representative integral membrane protein from E. coli. This protocol focuses on the application of fluorescence spectroscopy to determine equilibrium populations of folded and unfolded states as a function of denaturant concentration. Experimental considerations for the preparation of synthetic lipid vesicles as well as key steps in the data analysis procedure are highlighted. This technique is versatile and may be pursued with different types of denaturant, including temperature and pH, as well as in various folding environments of lipids and micelles. The current protocol is one that can be generalized to any membrane or soluble protein that meets the set of criteria discussed below.
Protokół
1. Preparation of ~50 nm Diameter Small Unilamellar Vesicles (SUVs) for Membrane Protein Folding
- A solution of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) lipids in chloroform is purchased and aliquotted into clean glass vials in 20 mg per vial quantities for storage. A layer of nitrogen gas is added to each vial to prevent lipid oxidation, and the vials are sealed with caps and parafilm. The vials are stored in a -20° C freezer until use.
- A single vial that contains an aliquot of 20 mg lipid in chloroform is used for each experiment. The contents of the vial are dried using a stream of nitrogen gas for 1 hour until no solvent remains.
- Dried lipids are resuspended in 1 mL of 20 mM potassium phosphate buffer (pH = 7.3) using a water bath sonicator for ~ 30 seconds. This solution of suspended lipid will appear cloudy, and is transferred to a plastic 15-mL tube with a conical bottom. Another 1 mL aliquot of phosphate buffer is added to the empty glass vial that originally contained lipid, and sonication is repeated; the solution is then added to the same 15-mL tube. This process is repeated a total of 4 times until the final volume in the tube is 4 mL, resulting in a lipid concentration of 5 mg/mL in this stock vesicle solution.
- The 4 mL volume of lipid vesicle solution is placed in a warm (~ 30° C) water bath, and sonicated using an ultrasonicator microtip for 1 hour at 50% duty cycle. The purpose of the water bath is twofold. First, it prevents the lipid solution from becoming too hot because of the sonication process. Second, the warm bath ensures that the temperature of the aqueous lipid solution remains above the bilayer phase transition temperature during sonication; for DMPC, this transition temperature is ~23° C.
- The sonicated solution is passed through a 0.22 μm syringe filter to remove debris from the sonicator tip, and this filtered solution is equilibrated overnight at 37° C.
2. Sample Preparation for Initial Fluorescence Unfolding Curve
- A stock solution of 10 M urea in 20 mM phosphate buffer (pH 7.3) is made. It is essential that water be added to the solid (and not vice versa) because the large quantity of urea occupies a significant volume in the solution. This urea solution can be made by adding solutes to a clean, empty bottle and adding water up to the intended final volume. This solution may require warming in a water bath or on a hot plate with stirring to dissolve the solute. Urea is hygroscopic and therefore, the actual concentration of the urea stock solution should be determined using refractive index.1
- A stock buffer solution of 20 mM phosphate (pH 7.3) is made.
- A stock solution of unfolded protein is prepared. This stock protein solution should have a concentration of ~200 μM protein in 8 M urea, 20 mM phosphate buffer.
- Samples for fluorescence studies are prepared in the following manner. Appropriate volumes of stock protein (section 2.3), stock lipid solution (5 mg/mL lipid, section 1), stock 10 M urea solution (section 2.1), and stock phosphate buffer (section 2.2) are combined to make samples containing ~4 μM protein, 1 mg/mL lipid, and 0 to 8 M urea in 1 M increments. The total volume of each sample is 200 μL. An example spreadsheet (Table 1) is included that lists volumes required using a 200 μM stock protein solution.
- Blank samples are made as described in section 2.4, however, protein is not added. In place of protein, 4 μL of an 8 M urea solution may be added so concentrations of other components are identical to those in section 2.4. These blanks are used to subtract scattering and other background signal that appears in the fluorescence spectra of protein.
- Samples from sections 2.4 and 2.5 are allowed to incubate at 37° C for at least 2 hours before measurement of fluorescence spectra to ensure complete folding and equilibration.
3. Measurement of Fluorescence Spectra
Tryptophan fluorescence of each sample and blank is measured using a steady-state fluorometer. Fluorescence spectra should be recorded with excitation wavelength of 290 nm to avoid excitation of tyrosine residues, and scanned from 305 to 500 nm. Typical entrance and exit bandpass is 3 nm. The wavelength increment and integration time can be optimized for signal-to-noise ratio. Typical values for wavelength increment and integration time are 1 nm/step and 0.5 sec/step, respectively. Membrane proteins generally fold into synthetic lipids only when the temperature is above the bilayer phase transition temperature. Therefore, in this case of DMPC, the temperature of the sample is held constant at 30 °C. A microvolume fused silica cuvette with 160 μL capacity is utilized in these experiments.
4. Generation of Initial Unfolding Curve and Estimation of Gibbs Free Energy of Membrane Protein Unfolding
- Fluorescence spectra in the form of an x-y dataset are loaded into software such as Igor Pro, Matlab, Origin, or Excel.
- Signal from the urea and lipid vesicle background must be subtracted using the following procedure:
Spectrum A = raw fluorescence spectrum of protein in the presence of lipid vesicles and specific concentration of urea.
Spectrum B = raw fluorescence spectrum of corresponding blank sample that contains lipid vesicles and same urea concentration as that in spectrum A; no protein is in the blank samples.
Corrected Spectrum = Spectrum A - C*spectrum B. C is a scalar typically equal to 1.0. In some cases, C is less than or greater than 1.0 because of scattering issues that cause over- or under-subtraction near the ~305-320 nm region. - The wavelength of maximum fluorescence, λmax, is tabulated from corrected spectra for each urea concentration. A typical value for fully unfolded membrane protein is 350 nm while that of folded protein is ~330 nm. Tabulated wavelengths are converted to fraction unfolded by converting the range of λmax values (~330 to ~350 nm) to the range 0.0 to 1.0 to correlate the λmax value to fraction of unfolded population. For example, a λmax value of 330 nm corresponds to 0.0, 350 nm corresponds to 1.0, and 340 nm corresponds to 0.5 in terms of fraction unfolded. This fraction of unfolded protein is then plotted against urea concentration. As stated above, the urea concentration should be determined experimentally by measuring refractive index.
- We assume that the protein can exist in one of two states: folded or unfolded. The plot of fraction unfolded, f, versus urea concentration, C, can be fit to the equation:2

The coefficient m corresponds to the rate of change of the free energy with respect to denaturant concentration, and Cm corresponds to the midpoint urea concentration at which the folded population equals the unfolded population. These values are the fitting variables obtained from least-squares fitting by data analysis software. R (0.001987 kcal/mol • K) is the gas law constant, and T is temperature (303 K). - The coefficients obtained from the fit are utilized to determine the Gibbs free energy (kcal/mol) of unfolding in the absence of urea:
 .
.
5. Optimized Unfolding Curve for More Precise Measurement of Gibbs Free Energy of Unfolding.
- The unfolding curve described above reveals the range of urea concentrations that causes the greatest change in the λmax. This range is typically small, and the majority of unfolding occurs within this small range. In order to precisely determine the free energy of unfolding, additional samples are analyzed in this region to obtain a plot that contains many data points necessary for optimized least-squares fitting. For example, if the protein unfolds between 2 and 4 M urea, samples can be made that contain 2 - 4 M urea in 0.2 M increments to more precisely reveal the shape of this important region in the unfolding curve. It is not necessary to measure blank spectra at each urea concentration; it is sufficient to measure a blank spectrum every ~0.5 M step.
- The free energy of unfolding obtained from this plot may differ slightly from that obtained in the original unfolding curve, but reflects a more precise value.
6. Representative Results:
| Sample | final [Urea], M | stock protein (μL) | stock lipid (μL) | stock 10 M urea (μL) | stock buffer (μL) |
| 1 | ~ 0.16 | 4 | 40 | 0 | 156 |
| 2 | 1 | 4 | 40 | 20 | 136 |
| 3 | 2 | 4 | 40 | 40 | 116 |
| 4 | 3 | 4 | 40 | 60 | 96 |
| 5 | 4 | 4 | 40 | 80 | 76 |
| 6 | 5 | 4 | 40 | 100 | 56 |
| 7 | 6 | 4 | 40 | 120 | 36 |
| 8 | 7 | 4 | 40 | 140 | 16 |
Table 1. Volume of stock solutions required to make fluorescence samples
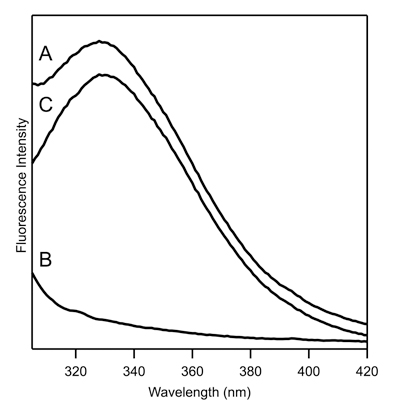
Figure 1. Tryptophan fluorescence spectra of ~ 5 μM representative membrane protein that contains a single tryptophan residue. (A) Raw fluorescence spectra of protein (spectrum A from 4.2); (B) Raw fluorescence spectra of blank (spectrum B from 4.2); (C) Corrected spectrum from 4.2.

Figure 2. Corrected tryptophan fluorescence spectra of membrane protein from Figure 1 for numerous urea concentrations.

Figure 3. Unfolding curve obtained from data in Figure 2, with fit to equation in 4.4. The Gibbs free energy is calculated according to section 4.5.
Dyskusje
The current protocol describes the generation of unfolding curves of membrane-associated proteins and peptides that contain tryptophan residues. Here, it is assumed that the tryptophan fluorescence reflects whether the protein is folded and inserted into synthetic lipid vesicles, or unfolded in solution. Additional assumptions, such as two-state folding and linear dependence of free energy with denaturant concentration, are made in the current report; modification of these assumptions result in different equations.
Ujawnienia
No conflicts of interest declared.
Podziękowania
We thank Beijing Wu for use of her data. This work was supported by an NSF CAREER award to J.E.K.
Materiały
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| Name | Company | Catalog Number | Comments | |
| DMPC | Avanti Polar Lipids | 850345C | ||
| Urea | MP Biochemicals | 04821527 | ||
| Potassium Phosphate Dibasic | Fisher | P288 | ||
| Potassium Phosphate Monobasic | Fisher | P285 |
Odniesienia
- Shirley, B. A., Shirley, B. A. . Protein Folding and Stability. , 177-190 (1995).
- Pace, C. N. Determination and Analysis of Urea and Guanidine Hydrochloride Denaturation Curves. Methods Enzymol. 131, 266-279 (1986).
- Lau, F. W., Bowie, J. U. A Method for Assessing the Stability of a Membrane Protein. Biochemistry. 36, 5884-5892 (1997).
- Burgess, N. K., Dao, T. P., Stanley, A. M., Fleming, K. G. β-Barrel Proteins that Reside in the Escherichia coli Outer Membrane in Vivo Demonstrate Varied Folding Behavior in Vitro. J. Biol. Chem. 283, 26748-26758 (2008).
- Booth, P. J., Curnow, P. Folding scene investigation: membrane proteins. Curr. Opin. Struct. Biol. 19, 8-13 (2009).
- Hong, H., Tamm, L. K. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. U.S.A. 101, 4065-4070 (2004).
- Sanchez, K. M., Gable, J. E., Schlamadinger, D. E., Kim, J. E. Effects of Tryptophan Microenvironment, Soluble Domain, and Vesicle Size on the Thermodynamics of Membrane Protein Folding: Lessons from the Transmembrane Protein OmpA. Biochemistry. 47, 12844-12852 (2008).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone