Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Porphyromonas gingivalis as a Model Organism for Assessing Interaction of Anaerobic Bacteria with Host Cells
W tym Artykule
Podsumowanie
This article presents two protocols: one to measure anaerobic bacteria that can successfully invade and survive within the host, and the other to visualize anaerobic bacteria interacting with host cells. This study can be applied to any cultivable anaerobe and any eukaryotic cell type.
Streszczenie
Anaerobic bacteria far outnumber aerobes in many human niches such as the gut, mouth, and vagina. Furthermore, anaerobic infections are common and frequently of indigenous origin. The ability of some anaerobic pathogens to invade human cells gives them adaptive measures to escape innate immunity as well as to modulate host cell behavior. However, ensuring that the anaerobic bacteria are live during experimental investigation of the events may pose challenges. Porphyromonas gingivalis, a Gram-negative anaerobe, is capable of invading a variety of eukaryotic non-phagocytic cells. This article outlines how to successfully culture and assess the ability of P. gingivalis to invade human umbilical vein endothelial cells (HUVECs). Two protocols were developed: one to measure bacteria that can successfully invade and survive within the host, and the other to visualize bacteria interacting with host cells. These techniques necessitate the use of an anaerobic chamber to supply P. gingivalis with an anaerobic environment for optimal growth.
The first protocol is based on the antibiotic protection assay, which is largely used to study the invasion of host cells by bacteria. However, the antibiotic protection assay is limited; only intracellular bacteria that are culturable following antibiotic treatment and host cell lysis are measured. To assess all bacteria interacting with host cells, both live and dead, we developed a protocol that uses fluorescent microscopy to examine host-pathogen interaction. Bacteria are fluorescently labeled with 2',7'-Bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein acetoxymethyl ester (BCECF-AM) and used to infect eukaryotic cells under anaerobic conditions. Following fixing with paraformaldehyde and permeabilization with 0.2% Triton X-100, host cells are labeled with TRITC phalloidin and DAPI to label the cell cytoskeleton and nucleus, respectively. Multiple images taken at different focal points (Z-stack) are obtained for temporal-spatial visualization of bacteria. Methods used in this study can be applied to any cultivable anaerobe and any eukaryotic cell type.
Wprowadzenie
Anaerobic bacteria colonize almost all surfaces of the human body. Although predominant in the flora of the intestinal and genitourinary tracts where oxygen concentrations are low, they also exist at high levels on the skin, mouth, nose, and throat1. Anaerobic bacteria are a common cause of endogenous infections and are frequently isolated from diseased sites. However, because of their fastidious nature, anaerobes can be difficult to isolate and culture. Studies involving anaerobic bacteria must be done under restricted conditions. Modern anaerobic-culture techniques allow researchers to mimic the anaerobic settings required to study many anaerobic laboratory strains or even clinical isolates2,3.
Pathogenic anaerobic bacteria have developed a dynamic relationship and co-evolution with the host cells in which they reside. Most anaerobes are susceptible to killing by the host immune response before reaching infectious levels. However, some pathogenic bacteria have developed mechanisms to escape from or subvert the host immune response. They accomplish this goal through mechanisms such as evasion of immune recognition, neutralization of immune mediators, alteration of cell-mediated immunity, invasion of host cells, and alteration of immune signaling4. Porphyromonas gingivalis, a Gram-negative anaerobe implicated in both oral and extraoral diseases, is one example of a highly adapted bacterial pathogen capable of causing pathogenic changes in the host5-7.
Pockets of biofilm plaque accrued in deep crevices formed between the teeth and gingival mucosal tissue can harbor anaerobic bacteria that are protected from atmospheric oxygen8. These periodontal pockets serve as a niche for various anaerobic pathogens, such as P. gingivalis9. P. gingivalis is a keystone pathogen that is capable of remodeling the oral microbial community in ways that promote development and progression of periodontal diseases10. It produces a large number of virulence factors that are active against a broad spectrum of host proteins and provides mechanisms for evasion of host defenses11. It is also capable of invading epithelial, endothelial, fibroblastic, and periodontal ligament cells in vitro12-14 and in vivo15. By effectively invading host cells, P. gingivalis can escape host immunity. Effective invasion of host cells not only allows the bacterium to escape host defenses but also serves as a reservoir for future re-infection as well as alters the host cell. Studies of the molecular mechanisms involved in adhesion and internalization of the bacterium by host cells are needed. Research in several laboratories is focused on understanding the molecular events associated with internalization of P. gingivalis by the host cells as well as the mechanisms used to suppress and hijack the immune response and survive the hostile host defense mechanisms.
There are many assays capable of identifying and characterizing pathogens that are capable of invading host cells. However, in vitro studies with anaerobic pathogens pose many experimental problems for the researcher mainly because it is difficult to perform studies that rely on bulky instruments in the absence of oxygen. This is compounded by the fact that eukaryotic cells require oxygen to grow and thus must be prepared separately in tissue culture incubators. One way to avoid such obstacles would be to perform the studies under atmospheric oxygen, but that would make growth of anaerobic bacteria impossible. Another method would be to use heat-killed bacteria to infect and study host-cell interactions. However, differences exist between heat-killed and viable bacteria that diminish the relevancy of the host-pathogen interaction16. It is central to study viable bacteria with unaltered expression interacting with host cells; therefore, methods for culturing P. gingivalis in an anaerobic setting are given. Also, two simple cost-effective protocols are demonstrated for assessing the ability of P. gingivalis to be internalized by human umbilical vein endothelial cells (HUVECs). The first protocol is based on the popular antibiotic protection assay. Although the assay is straightforward, considerations when using anaerobic microorganisms are given. The second protocol requires the use of a fluorescent scanning microscope to visualize interacting and internalized P. gingivalis. Each assay has its limitations and advantages that will be discussed to provide the researcher an outline for studying the invasiveness of anaerobic bacteria. Although the current manuscript studies P. gingivalis and HUVECs, these protocols can be used for many other anaerobic bacteria as well as for other types of host cells.
Protokół
The following protocols will describe methods for culturing and studying the invasion by the anaerobic species, P. gingivalis; however, these protocols may be used for a number of anaerobic pathogens. Although HUVECs are used, this protocol may be used for other eukaryotic cells both immune and non-immune.
1. Anaerobic Chamber Use and Maintenance
Note: P. gingivalis is an anaerobe sensitive to normal levels of oxygen encountered in ambient air. A controlled anaerobic environment is vital for the cultivation of P. gingivalis.
- Here, maintain an artificial atmosphere designated as mixed anaerobic gas (80% N2, 10% H2, 10% CO2) in a vinyl anaerobic chamber (Figure 1A). Use an airlock (Figure 1B) for transferring items from the laboratory environment to the anaerobic chamber. The airlock operates manually, twice purging with N2 gas before introducing the mixed anaerobic gas.
- Use a hydrogen sulfide removal column (Figure 1C) for maintenance-free removal of the undesirable hydrogen sulfide. Place a dehumidifier within the chamber to remove H2O created by the catalyst and to avoid aerosols that facilitate the spread of contamination.
Note: Hydrogen sulfide is a natural metabolic byproduct of many anaerobic bacteria and its accumulation is toxic to bacteria and can result in damage to electronics and decrease the lifetime of a catalyst. - Use a fan box to circulate the chamber’s atmosphere through a palladium catalyst, which removes oxygen in the presence of hydrogen (Figure 1D).
Note: A recirculating atmospheric (HEPA) filter removes airborne contaminants with a size of 0.22 µm or larger. - Culture anaerobic bacteria in a 37 °C incubator that is located inside of the anaerobic chamber. Use standard aseptic techniques when working inside the anaerobic chamber.
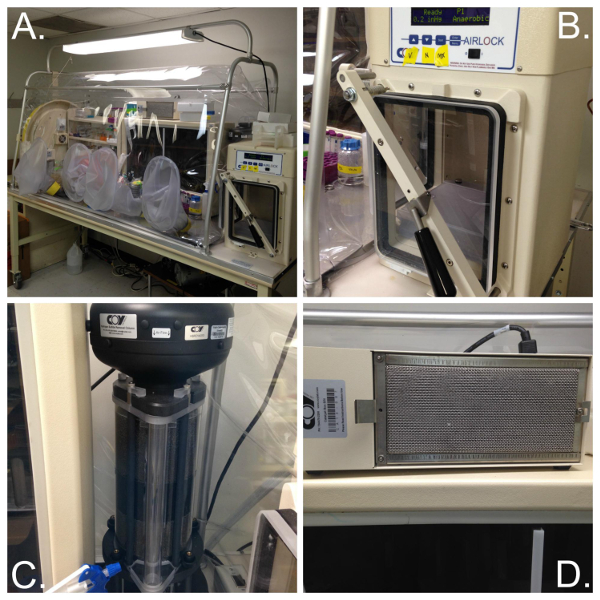
Figure 1. Anaerobic vinyl chamber and its components. (A) A vinyl anaerobic chamber sealed completely from atmospheric oxygen provides workspace for two individuals at a time (32 in x 78 in). It contains an incubator set at 37 °C (back middle). (B) An airlock is used for the transfer of items from the lab environment to the anaerobic chamber. Pictured is an automatic airlock operated through a controller that can be programmed to automatically perform the vacuum and purge procedures needed to create an anaerobic environment. (C) A Hydrogen Sulfide Removal Column provides maintenance-free high capacity removal of undesirable hydrogen sulfide. (D) Two catalyst fan boxes are placed throughout the anaerobic chamber to help circulate the chamber’s atmosphere through palladium catalyst, which, in the presence of hydrogen, removes oxygen. The anaerobic chamber is set up according to manufacturer’s instructions. Please click here to view a larger version of this figure.
2. Preparation of Anaerobic Bacteria
Note: P. gingivalis is aerotolerant and can be stored in aerobic conditions but it will not grow in the presence of oxygen at levels higher than 6%17,18. An anaerobic chamber is necessary for the proper cultivation of P. gingivalis and other anaerobic species (Figure 1). Proper training and education on anaerobic chamber use is required before working with microanaerobes19.
- Equilibrate all liquid media and plates to anaerobic conditions for at least 12 hr prior to experimentation to remove residual oxygen.
- Transfer P. gingivalis from -80 °C freezer to anaerobic chamber, let thaw.
- Streak P. gingivalis on trypticase soy blood agar plates (TSA II with 5% sheep blood). Wrap plates in parafilm and store at 37 °C in an anaerobic incubator for 4-7 days.
- Inoculate P. gingivalis into 3 ml brain heart infusion (BHI) broth supplemented with hemin and menadione, an enriched non-selective liquid media for the isolation and culture of anaerobic and fastidious microorganisms, using sterile loops.
Note: For long-term storage, mix bacterial cultures prepared in BHI with glycerol or DMSO (10-20% final concentration) and place in a -80 °C freezer. - Prepare a starter culture of P. gingivalis by making a 1:10 dilution and allowing bacteria to grow until mid-log phase.
Note: The optical density of the bacterial suspension is determined and the bacterial concentration for each strain to be examined is adjusted. For P. gingivalis a suspension at OD660 of 0.7 corresponds to mid-log phase and ~7 x 108 cells/ml. Growth conditions described in the protocol above are specific for P. gingivalis and may need to be adapted for other bacterial strains.
3. Endothelial Cell Culture
Note: Purchase pooled primary HUVECs and culture in basal medium containing vascular endothelial growth factors (VEGF) at 37 °C in 5% CO2 according to manufacturer's instructions.
- Seed HUVECs in T-75 flasks at 2.5 x 105 cells/flask in 15 ml VEGF media.
Note: Check viability via a 1:1 dilution with 4.0% trypan blue. Cells with a compromised membrane will retain trypan blue, healthy cells with intact membranes will appear white when viewed under a binocular light microscope. Count 100 cells, ensure that over 80% of the cells are viable20. - Replace media every 2 days with pre-warmed fresh VEGF media until cells reach ~80% confluency.
- Wash cells once with pre-warmed PBS. Liberate cells from the T75 flask by incubating with 2 ml trypsin-EDTA (0.25%) for 5 min followed by 2 ml trypsin neutralizing solution.
- Collect suspended HUVECs in a 50 ml conical tube. Wash out any extra cells from T-75 flasks with PBS and transfer to 50 ml conical tubes.
- Centrifuge cells at 200 x g for 10 min.
- Remove supernatant, suspend cell pellet in 10 ml pre-warmed VEGF media.
- Determine cell concentration using a hemocytometer or similar cell counting device.
- Calculate amount of cell suspension to add to either 6-well plate (400,000/well) or 12-well plate with coverslips (50,000/well). HUVECs will be ready for experimentation the next day.
4. Survival Assay Invasion/Interaction (Plating)
Note: When performing this assay, prepare two 6-well plates of endothelial cells seeded at 400,000 cells/well. One plate will be used to assess bacteria attached to and internalized by host cells. The other plate will account for intracellular bacteria. The 6-well plate allows for triplicates of two samples to be performed in one experiment. For an outline of this protocol please refer to the survival assay flowchart (Figure 2).
- Prepare anaerobic bacteria as described above (see section 1) until they reach mid-log growth (OD660 0.5-0.7).
- Centrifuge bacteria at 5,000 x g for 10 min.
Note: If centrifuge is outside anaerobic chamber, carry bacterial samples in a tightly sealed 15 ml tube, wrap the cap with parafilm to prevent oxygen leakage. - Place pelleted P. gingivalis back in chamber, discard supernatant. Wash with PBS, pellet bacteria again before resuspending in VEGF media. Prepare suspensions for all bacterial strains to be tested at OD660 of 0.7 which corresponds to mid-log phase (~7 x 108 cells/ml). The bacteria are now ready for infection.
- Transfer 6-well plates containing HUVECs from tissue culture incubator into anaerobic chamber. Remove media and wash three times with anaerobic PBS. Add 2 ml of anaerobic VEGF media to each well and place the plates at 37 °C in the anaerobic incubator for 20 min to equilibrate the temperature for infection.
Note: Plate bacteria on blood agar plates to ensure the ones used for infection are homogenous and not contaminated upon infection. - Infect host cells with bacteria at a multiplicity of infection (MOI bacteria: host) of 100:1.
Note: HUVEC cell number is determined by performing a trypan exclusion test on a single well before infection. Bacterial cell number is determined via optical density (e.g., OD of 0.5 = 5 x 108 cells/ml). Bacterial concentration is adjusted to proper MOI based on HUVECs concentration21. - Place 6-well plates with infected HUVECs into anaerobic incubator and allow bacteria to interact with host cells for 30 min.
- Prepare saponin in BHI (1.0% w/v) inside the anaerobic chamber and filter through a 0.2 µm filter.
- Survival of both attached and internalized bacteria.
- Remove plates from the incubator, aspirate media, wash three times with anaerobic PBS and add 2 ml filtered 1.0% saponin (prepared as described in step 4.8). Incubate for 15 min to allow host cell lysis.
- Scrape bottom of each well with cell scraper. Collect the cell mixture from each well and make a 1:1 dilution in BHI.
- Proceed to make serial dilutions of the sample. Depending on bacterial species and concentration, adjust serial dilutions. Start with 1:100 or 1:1,000 dilutions.
- Plate 200 µl of desired dilution on blood agar plates. Wrap the plates in parafilm and place in the anaerobic incubator at 37 °C.
- Following seven days of incubation at 37 °C, remove the plates and count colony forming units (CFUs) using a light box to manually count colonies.
Note: CFUs are enumerated. For larger quantities of CFUs, images may be taken and computer software can be used to facilitate the enumeration of CFUs.
- Survival of internalized bacteria.
- Remove plates from the incubator. Wash three times with anaerobic PBS and add 2 ml VEGF media with supplemented antibiotic (300 µg/ml of gentamicin and 400 µg/ml of metronidazole).
- Incubate for 1 hr. Be sure to test the antibiotics so they are 100% effective at killing the desired bacterial strain and make sure that they do not penetrate host cells22,23.
- Aspirate media, add 2 ml of filtered 1.0% saponin. Incubate for 15 min to allow host cell lysis.
- Scrape bottom of each well with cell scraper. Collect the cell mixture from each well and make 1:1 dilution in BHI.
- Prepare serial dilutions of the sample (1:100, 1:1,000).
- Plate 200 µl of desired dilution on blood agar plates. Wrap plates in parafilm and place in anaerobic incubator.
- After seven days of incubation at 37 °C remove plates and count CFUs.

Figure 2. Schematic representation of a protocol used for survival of anaerobic bacteria with eukaryotic cells. Both assays for a total bacterial survival and survival of internalized bacteria can be performed at the same time. Please click here to view a larger version of this figure.
5. Internalization of Bacteria into Host Cells (Fluorescent Microscopy)
Note: P. gingivalis is labeled with 2',7'-Bis-(2-Carboxyethyl)-5-(and-6)-Carboxyfluorescein, Acetoxymethyl Ester (BCECF-AM). BCECF-AM is a non-fluorescent membrane-permeable dye; its conversion to fluorescein BCECF via the action of intracellular esterases can indicate cell viability. P. gingivalis is labeled with the BCECF-AM dye and then used to infect eukaryotic cells. Following infection, cells are fixed and labeled with DAPI and TRITC-phalloidin. The DAPI stain used to stain the eukaryotic cell nucleus will also label bacterial cell nucleus, which provides a counter-measure to identify non-viable bacteria that cannot metabolically cleave BCECF-AM. Host cells are highlighted with TRITC-phalloidin, a red actin dye.
- Autoclave coverslips. Aseptically add coverslips to 12-well plates before seeding endothelial cells at 5 x 104 cells/well. (Prepared day before experiment)
- Have endothelial cells prepared on 18 mm (#1.5 thickness) circular coverslips in 12-well plates as described above.
- Prepare anaerobic bacteria grown to mid-log phase (OD660 = 0.5-0.7) as described in section 1.
- Wash bacteria 2x with anaerobic PBS by centrifuging at 5,000 x g and suspending pellet in PBS at 5-7 x 108 cells/ml.
- Add 20 µl of 0.2 mM BCECF-AM to 2 ml of bacterial suspension (5-7 x 108 cells/ml) to a final concentration of BCECF-AM of 2 µM.
- Incubate at 37 °C for 30 min in the dark.
- Transfer plates with endothelial cells seeded on 18 mm (#1.5 thickness) circular coverslips from tissue culture incubator into the anaerobic chamber. Wash with PBS and exchange with anaerobic VEGF media.
Note: Verify that HUVECs are healthy under a light microscope. HUVECS should be ~80% confluent, morphology should be comparable to the manufacturers. - Centrifuge labeled bacteria at 5,000 x g for 10 min to remove residual BCECF-AM dye. Suspend in 2 ml anaerobic VEGF media.
- Infect host cells with labeled bacteria at MOI of 100:1 (bacteria: host).
- Incubate in anaerobic chamber at 37 °C for 30 min.
- After infection wash cells with PBS three times and fix in freshly prepared 4.0% paraformaldehyde for 10 min.
Note: After fixing cells, experiment can be conducted outside of the anaerobic chamber. - Wash coverslips with PBS three times.
- Add 1 ml of 0.2% Triton X-100 for 10 min.
- Wash coverslips with PBS three times.
- Add 50 µl of TRITC phalloidin (50 µg/ml) to coverslips for 45 min.
- Wash coverslips three times, remove from the 12-well plate and place on a slide with soft-set mounting medium containing DAPI. Seal the sides with nail polish.
Note: Slides may be stored for a couple months in the dark. Avoid light exposure to prevent photo-bleaching. - View slides using a confocal microscope.
- Here, use a 34 channel spectral system (32-channel array detector and two side PMT detectors, plus a transmitted light detector) configured around an AxioObserver (inverted) stand with a motorized XY stage. The system has five lasers: blue diode (405 nm), multi-line Argon (458, 488, 514 nm), green diode (561 nm), red HeNe (633 nm) and a 440 nm pulsed laser. Equip a Fluorescence Lifetime Imaging system with 2 hybrid GaAsP detectors (for FRET-FLIM).
- Detect fluorescence from DAPI and TRITC in one channel using a dual-band filter with excitation wavelengths of 340-380 nm and 540-560 nm, and an emission filter of 435-485 nm and 570-590 nm respectively. Detect fluorescence from BCECF-AM using a filter with excitation wavelength of 440-500 nm and an emission filter of 510-590 nm.
Note: Controls for BCECF-AM should be done on every bacterial strain being studied to ensure proper labeling of viable bacteria. First validate that nonviable bacteria are DAPI-positive and BCECF-negative. Second, ensure that live bacteria can metabolize BCECF-AM into fluorescein BCECF. Variable concentrations of the bacteria or BCECF-AM dye may need to be tested for optimal labeling.
Wyniki
Protocols outlined above were used in studying host-pathogen interaction between P. gingivalis and endothelial cells. P. gingivalis W83 and a P. gingivalis V3150 carrying a deletion of PG0228 were used in the study. The PG0228 is predicted to encode a protein that may alter the levels of RNA and proteins, which may ultimately affect interaction of P. gingivalis with host cells. To investigate the effect of PG0228 on P. gingivalis’s ability to interact with host cells, the ab...
Dyskusje
All the above methods can be used to design specific assays to assess the interaction of anaerobic bacteria with eukaryotic cells. However, there are several considerations to successfully perform the experiments. First are the microbial strains to be used in a study.
It is crucial in the comparison of two strains with both the survival assay as well as by microscopy analysis that they are at similar growth phases and attain similar cell concentrations as any differences in the above can influ...
Ujawnienia
Authors have nothing to disclose.
Podziękowania
We would like to thank Dr. Hiroshi Miyazaki, Dr. Scott Henderson, Dr. Todd Kitten, Dr. Justin Hutcherson, Dr. Catherine Jauregui, and Collin R. Berry. This work was supported by NIH NIDCR grants R01DE016124, R01DE018039, and R01DE023304 to J.P. Lewis.
Microscopy was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility, supported, in part, with funding from NIH-NINDS Center core grant (5P30NS047463).
Materiały
| Name | Company | Catalog Number | Comments |
| Vinyl Anaerobic Chamber-Type B | Coy Laboratory Products | Model 2000 incubator | |
| TSA II Trypticase Soy Agar with 5% Sheep Blood | BBL | 221261 | |
| Human Umbilical Vein Endothelial Cells 10-donor Pool | LifeLine Technology | FC-0044 | |
| VascuLife VEGF Medium Complete Kit | LifeLine Technology | LL-0003 | |
| TrypKit | LifeLine | LL-0013 | |
| Saponin | Riedel-de Haen | 16109 | |
| Gentamicin Sulfate Salt | Sigma-Aldrich | G-1264 | |
| Metronidazole | Sigma-Aldrich | M-3761 | |
| BCECF-AM | LifeTechnologies | B1150 | |
| TRITC Phalloidin | Sigma-Aldrich | P1951 | |
| 18 mm Circular Coverslips | Electron Microscopy Sciences | 72222-01 | |
| VectaShield Mounting Medium with DAPI | Vector Laboratories | H-1200 |
Odniesienia
- Hentges, D. J. The Anaerobic Microflora of the Human Body. Clin. Infect. Dis. 16 (4), S175-S180 (1993).
- Willis, A. T. . Anaerobic bacteriology: clinical and laboratory practice. , (2014).
- Wren, M. W., Baldwin, A. W., Eldon, C. P., Sanderson, P. J. The anaerobic culture of clinical specimens: a 14-month study. J. Med. Microbiol. 10 (1), 49-61 (1977).
- Woolard, M. D., Frelinger, J. A. Outsmarting the host: bacteria modulating the immune response. Immunol. Res. 41 (3), 188-202 (2008).
- Mayrand, D., Holt, S. C. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol. Rev. 52 (1), 134-152 (1988).
- Lamont, R. J., Jenkinson, H. F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62 (4), 1244-1263 (1998).
- Haffajee, A. D., Socransky, S. S. Microbial etiological agents of destructive periodontal diseases. Periodontol. 2000. 5 (1), 78-111 (1994).
- Listgarten, M. A. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J. Periodontol. 47 (1), 1-18 (1976).
- Ximénez-Fyvie, L. A., Haffajee, A. D., Socransky, S. S. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27 (9), 648-657 (2000).
- Darveau, R. P., Hajishengallis, G., Curtis, M. A. Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 91 (9), 816-820 (2012).
- Holt, S. C., Kesavalu, L., Walker, S., Genco, C. A. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000. 20 (1), 168-238 (1999).
- Lamont, R. J., Yilmaz, &. #. 2. 4. 6. ;. Z. In or out: the invasiveness of oral bacteria. Periodontol. 2000. 30 (1), 61-69 (2002).
- Lamont, R. J., et al. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63 (10), 3878-3885 (1995).
- Belton, C. M., Izutsu, K. T., Goodwin, P. C., Park, Y., Lamont, R. J. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell. Microbiol. 1 (3), 215-223 (1999).
- Rautemaa, R., et al. Intracellular localization of Porphyromonas gingivalis thiol proteinase in periodontal tissues of chronic periodontitis patients. Oral Dis. 10 (5), 298-305 (2004).
- Kaufmann, S. H. Immunity to intracellular bacteria. Annu. Rev. Immunol. 11 (1), 129-163 (1993).
- Diaz, P., Rogers, A. The effect of oxygen on the growth and physiology of Porphyromonas gingivalis. Oral Microbiol. Immunol. 19 (2), 88-94 (2004).
- Lewis, J. P., Iyer, D., Anaya-Bergman, C. Adaptation of Porphyromonas gingivalis to microaerophilic conditions involves increased consumption of formate and reduced utilization of lactate. Microbiology. 155, 3758-3774 (2009).
- Edwards, A. N., Suarez, J. M., McBride, S. M. Culturing and maintaining Clostridium difficile in an anaerobic environment. J. Vis. Exp. (79), e50787 (2013).
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. , (2001).
- Koch, A. L., Crandall, M. Photometric measurement of bacterial growth. The American Biology Teacher. 30 (6), 481-485 (1968).
- Wikins, T. D., Holdeman, L. V., Abramson, I. J., Moore, W. E. Standardized single-disc method for antibiotic susceptibility testing of anaerobic bacteria Antimicrob. Agents Chemother. 1 (6), 451-459 (1972).
- Bauer, A. W., Kirby, W. M., Sherris, J. C., Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45 (4), 493-496 (1966).
- Mandell, G. L. Interaction of intraleukocytic bacteria and antibiotics. J. Clin. Invest. 52 (7), 1673-1679 (1973).
- Menzies, B. E., Kourteva, I. Internalization of Staphylococcus aureus by endothelial cells induces apoptosis. Infect. Immun. 66 (12), 5994-5998 (1998).
- Naito, M., et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 15 (4), 215-225 (2008).
- Goebel, W., Kuhn, M. Bacterial replication in the host cell cytosol. Curr. Opin. Microbiol. 3 (1), 49-53 (2000).
- Gospodarowicz, D. C. Extracellular matrix and control of proliferation of vascular endothelial cells. J. Clin. Invest. 65 (6), 1351-1364 (1980).
- DeQuach, J. A., et al. Simple and high yielding method for preparing tissue specific extracellular matrix coatings for cell culture. PloS One. 5 (9), e13039 (2010).
- Sellers, J. R., Cook, S., Goldmacher, V. S. A cytotoxicity assay utilizing a fluorescent dye that determines accurate surviving fractions of cells. J. Immunol. Methods. 172 (2), 255-264 (1994).
- Van Veen, H. W., et al. Generation of a proton motive force by the excretion of metal-phosphate in the polyphosphate-accumulating Acinetobacter johnsonii strain 210A. J. Biol. Chem. 269 (47), 29509-29514 (1994).
- Jackson, V. N., Halestrap, A. P. The kinetics, substrate, and inhibitor specificity of the monocarboxylate (lactate) transporter of rat liver cells determined using the fluorescent intracellular pH indicator, 2',7'-bis(carboxyethyl)-5(6)-carboxyfluorescein. J. Biol. Chem. 271 (2), 861-868 (1996).
- He, J., et al. Role of Porphyromonas gingivalis FeoB2 in metal uptake and oxidative stress protection. Infect. Immun. 74 (7), 4214-4223 (2006).
- Anaya-Bergman, C., et al. Porphyromonas gingivalis ferrous iron transporter FeoB1 influences sensitivity to oxidative stress. Infect. Immun. 78 (2), 688-696 (2010).
- Ueshima, J., et al. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect. Immun. 71 (3), 1170-1178 (2003).
- Johnson, M. B., Criss, A. K. Fluorescence microscopy methods for determining the viability of bacteria in association with mammalian cells. JoVE. (79), (2013).
- Cordes, T., Maiser, A., Steinhauer, C., Schermelleh, L., Tinnefeld, P. Mechanisms and advancement of antifading agents for fluorescence microscopy and single-molecule spectroscopy. Physical Chemistry Chemical Physics. 13 (14), 6699-6709 (2011).
- Pawley, J. . Handbook of biological confocal microscopy. , (2010).
- Gursoy, U., Könönen, E., Uitto, V. Prevotella intermedia ATCC 25611 targets host cell lamellipodia in epithelial cell adhesion and invasion. Oral Microbiol. Immunol. 24 (4), 304-309 (2009).
- Sengupta, D., et al. Interaction of Prevotella intermedia strain 17 leucine-rich repeat domain protein AdpF with eukaryotic cells promotes bacterial internalization. Infect. Immun. 82 (6), 2637-2648 (2014).
- Reyes, L., Herrera, D., Kozarov, E., Roldán, S., Progulske-Fox, A. Periodontal bacterial invasion and infection: contribution to atherosclerotic pathology. J. Clin. Periodontol. 40, S30-S50 (2013).
- Grant, M. M., et al. Oxygen tension modulates the cytokine response of oral epithelium to periodontal bacteria. J. Clin. Periodontol. 37 (12), 1039-1048 (2010).
- Biedermann, A., Kriebel, K., Kreikemeyer, B., Lang, H. Interactions of Anaerobic Bacteria with Dental Stem Cells: An In Vitro Study. PloS One. 9 (11), e110616 (2014).
- Kriebel, K., Biedermann, A., Kreikemeyer, B., Lang, H. Anaerobic Co-Culture of Mesenchymal Stem Cells and Anaerobic Pathogens-A New In Vitro Model System. PloS One. 8 (11), e78226 (2013).
- Peyyala, R., Kirakodu, S. S., Novak, K. F., Ebersole, J. L. Oral microbial biofilm stimulation of epithelial cell responses. Cytokine. 58 (1), 65-72 (2012).
- Halldorsson, S., Lucumi, E., Gòmez-Sjöberg, R., Fleming, R. M. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens Bioelectro. 63, 218-231 (2015).
- Iyer, D., et al. AdpC is a Prevotella intermedia 17 leucine-rich repeat internalin-like protein. Infect. Immun. 78 (6), 2385-2396 (2010).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone