Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Whole-body Mass Spectrometry Imaging by Infrared Matrix-assisted Laser Desorption Electrospray Ionization (IR-MALDESI)
W tym Artykule
Podsumowanie
A mass spectrometry imaging (MSI) source operated at atmospheric pressure was developed by coupling mid-infrared laser desorption and electrospray post-ionization. Exogenous ice matrix was used as the energy-absorbing matrix to facilitate resonant desorption of tissue-related material. This manuscript provides a step-by-step protocol for performing IR-MALDESI MSI of whole-body neonatal mouse.
Streszczenie
Ambient ionization sources for mass spectrometry (MS) have been the subject of much interest in the past decade. Matrix-assisted laser desorption electrospray ionization (MALDESI) is an example of such methods, where features of matrix-assisted laser desorption/ionization (MALDI) (e.g., pulsed nature of desorption) and electrospray ionization (ESI) (e.g., soft-ionization) are combined. One of the major advantages of MALDESI is its inherent versatility. In MALDESI experiments, an ultraviolet (UV) or infrared (IR) laser can be used to resonantly excite an endogenous or exogenous matrix. The choice of matrix is not analyte dependent, and depends solely on the laser wavelength used for excitation. In IR-MALDESI experiments, a thin layer of ice is deposited on the sample surface as an energy-absorbing matrix. The IR-MALDESI source geometry has been optimized using statistical design of experiments (DOE) for analysis of liquid samples as well as biological tissue specimens. Furthermore, a robust IR-MALDESI imaging source has been developed, where a tunable mid-IR laser is synchronized with a computer controlled XY translational stage and a high resolving power mass spectrometer. A custom graphical user interface (GUI) allows user selection of the repetition rate of the laser, number of shots per voxel, step-size of the sample stage, and the delay between the desorption and scan events for the source. IR-MALDESI has been used in variety of applications such as forensic analysis of fibers and dyes and MSI of biological tissue sections. Distribution of different analytes ranging from endogenous metabolites to exogenous xenobiotics within tissue sections can be measured and quantified using this technique. The protocol presented in this manuscript describes major steps necessary for IR-MALDESI MSI of whole-body tissue sections.
Wprowadzenie
Mass spectrometry imaging (MSI) in microprobe mode involves desorption of the sample from a surface by a beam (laser or ions) at discrete locations over the surface of a sample. At each raster point, a mass spectrum is generated and the acquired spectra, along with the spatial location from which they were collected, can be used to simultaneously map numerous analytes within the sample. This label-free manner of imaging coupled to the sensitivity and specificity of mass spectrometry have helped MSI become one of the most rapidly evolving fields in mass spectrometry1,2.
Matrix-assisted laser desorption/ionization (MALDI) is the most common ionization method used for MSI analyses. However, the need for an organic matrix and the vacuum requirements of MALDI pose significant limitations on reproducibility, sample throughput, and the types of samples that can be analyzed using the method. A number of atmospheric pressure (AP) ionization methods have been developed in recent years to circumvent these restrictions3. These ambient ionization methods allow for analysis of biological samples in an environment that is much closer to their natural state and simplify sample preparation steps prior to analysis. Matrix-assisted laser desorption electrospray ionization (MALDESI) is an example of such an ionization method4,5.
In IR-MALDESI experiments, a thin layer of ice is deposited on the tissue surface as the energy-absorbing matrix. A mid-IR laser pulse is absorbed by the ice matrix, and facilitates desorption of neutral materials from the surface by resonantly exciting the O-H stretching mode of water. The desorbed neutrals partition into the charged droplets of an orthogonal electrospray and are post-ionized in an ESI-like fashion4-6. The addition of exogenous ice matrix is preferred over relying solely on the endogenous water in tissue since it helps account for variations in water content in different tissue compartments, and has been shown to enhance desorption6 and improve ion abundance by ~15-fold7,8 in tissue imaging experiments.
In this work, we utilize IR-MALDESI MSI to elicit the distribution of metabolites across different organs in a neonatal mouse whole body. An overview of adjustable parameters of the IR-MALDESI source is given, and the necessary steps for successful imaging of tissue sections are demonstrated.
Protokół
Note: The following protocol describes all the necessary steps for performing IR-MALDESI MSI experiments. In-depth details about the optimized geometry of the IR-MALDESI source and its synchronization with the laser, stage, and mass spectrometer can be found elsewhere5,6. Animal tissue samples used in this protocol were obtained according to Institutional Animal Care and Use Committee (IACUC) and North Carolina State University regulations.
1. Tissue Preparation
- Prepare an isopentane/dry ice bath by placing ~200 ml of isopentane in a clean beaker inside a secondary container of dry ice in a fume hood. Use protective gloves and safety goggles at all times when handling the isopentane/dry ice bath and the tissue.
- Euthanize 2-day-old whole neonatal mouse pup by Avertin overdose (7.5 mg/g body weight), and then freeze the tissue in the isopentane/dry ice bath to preserve tissue structure. Use a pre-cleaned pair of forceps to place and remove the mouse pup in cryomold from the isopentane/dry ice bath. Store the flash-frozen tissue at -80 °C until analysis.
- Using protective gloves, apply a layer of optimal cutting temperature (OCT) mounting medium to a 40 mm cryostat specimen disc. Gently place the frozen whole mouse onto the OCT coated specimen holder to adhere mouse to the disc in the desired orientation for sectioning.
Note: OCT is used solely to adhere the tissue to the specimen disc for sectioning. Do not completely embed tissue in OCT and avoid excess as OCT is known to have detrimental effects on MSI analysis. Other media, such as gelatin, can be used to cryo-embed the tissue before slicing it9. - Place the disc (with OCT and tissue section) on Peltier sample holder in cryostat and press Peltier power button. Wait 10 min for sample to come to thermal equilibrium.
- Place the specimens disc (with the tissue mounted on it) inside the disc holder, and face the tissue down to the desired plane for analysis. Slice the tissue into sections of the desired thickness at -20 °C using the rotary microtome housed inside the cryostat10.
Note: For whole-body analyses, slice the tissue into 25-μm-thick sections to maintain the tissue integrity. For smaller regions (e.g., liver, brain, kidney), slice the tissue into 10-μm-thick sections.- Use the anti-roll glass plate to prevent the sliced tissue section from rolling. Use the cryostat vacuum hose and brushes to remove unwanted tissue debris. Once the tissue is sectioned, use the blade guard to cover the sharp microtome blade to avoid personal injuries.
- Orient mouse section on the specimen plate and thaw-mount onto a pre-cleaned glass microscope slide by bringing the slide as close as possible to the tissue section, without touching it10.
Note: For quantitative MSI experiments, coat the slide with an internal standard using an automated pneumatic sprayer prior to mounting the tissue, and thaw-mount the tissue section on the coated slide11. - Remove the microtome blade used to sectioning the tissue using the blade ejector, and safely discard the blade in the appropriate "sharp waste" container.
- Keep the glass slide inside the cryostat until Step 3.2 to maintain cryopreservation of the tissue.
2. IR-MALDESI Preparation/Calibration
- Turn on the mid-IR laser and launch the laser control application on the computer. Choose the desired wavelength using the laser control application supplied by the manufacturer. IR-MALDESI experiments are typically performed at 2,940 nm.
Note: Recent experiments have investigated the dependence of mid-IR laser wavelengths and the ice matrix with noted differences which appear to be analyte specific8. - Turn on the stage controller, launch the custom user control program (RASTIR), and calibrate the stage position using the home button.
- Prepare electrospray solution. Typically, use a 50:50 (v/v) methanol/water with 0.2% formic acid for the electrospray solvent in positive-ion mode. Select the electrospray solvent composition according to the experiment. For instance, use 5 mM ammonium hydroxide as electrospray modifier for imaging applications in negative-ion mode.
Note: For polarity switching IR-MALDESI MSI, use 1 mM acetic acid as the modifier. This modifier was found optimum for obtaining stable and reproducible signal in both positive- and negative-ion modes12. - Fill a 1 ml syringe with electrospray solvent, and flush the silica capillary with new solvent.
- Align ESI emitter on axis with MS inlet using the source parameters such as ESI-spot distance, spot-inlet distance, sample height, and solvent flow rate that have been optimized using statistical DOE for analysis of tissue sections6. Note: See Figure 1 for a schematic depicting these parameters and optimized values for MSI of tissue sections.
- Start the electrospray and evaluate its stability (>10 min) by monitoring the total ion current (TIC) which, at this point, consists solely of ambient compounds. Typically, a TIC variation of <10% over 10-15 min is a good indication of stability.
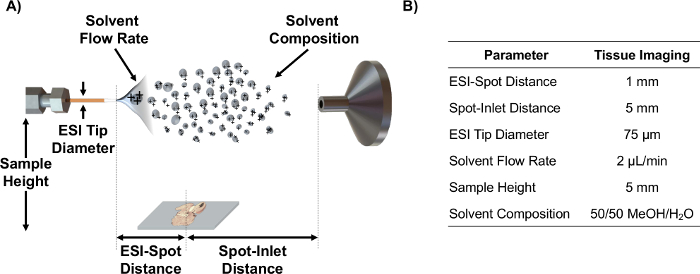
Figure 1. IR-MALDESI schematic and parameters. (A) Schematic of IR-MALDESI source setup (not to scale) and the adjustable parameters. (B) Optimized parameter values for imaging of tissue sections. Please click here to view a larger version of this figure.
3. Deposition of Ice Matrix
- Caution: Turn off the electrospray before placing sample on the stage.
- Place the thaw-mounted tissue onto the IR-MALDESI sample plate within the camera view.
- Make sure hands are clear of the ESI emitter and restart the electrospray.
- Close the access door of the IR-MALDESI source and purge enclosure with dry nitrogen to prevent condensation of water on the surface of the tissue. Once the relative humidity inside the enclosure reaches <3%, turn on the DC power supply (~12 V) to the Peltier plate, which will cool the sample plate. Note: The cooling process in the Peltier-cooled stage is discussed in detail elsewhere13.
- Cool the stage to -9 °C, and allow the mounted tissue to come to thermal equilibrium (~5-10 min).
- Stop nitrogen purge and expose the tissue to the ambient relative humidity by opening the source access door. A thin layer of ice will be deposited onto the cold surface of the Peltier stage and mounted tissue section due to desublimation of water from the air.
Note: If the relative humidity in the lab is below 10-15%, place a beaker of warm water inside the enclosure to facilitate the formation of the ice matrix layer. - After the ice matrix layer is formed, close the access door and restart the flow of dry nitrogen to reduce the relative humidity to 10±2%. Adjust the nitrogen flow rate to keep the relative humidity at this level, empirically found to be the balance of ice formation and sublimation to maintain a constant layer of ice throughout the experiment6.
4. Mass Spectrometry Imaging Data Acquisition
- Using the RASTIR GUI (Figure 2), move the stage to the analysis position by clicking the "LASER position" button. Use the adjustment diode of the laser to ensure an off-tissue region will be ablated. Fire a laser shot in an off-tissue region to calibrate the laser offset with respect to camera view. Click the "Laser Test Fire" button to update the laser position.
- Return to "CAMERA position" and update the laser position in RASTIR by placing the laser alignment reticle on the laser ablation spot (Figure 2-1).
- Click the region of interest (ROI) button and adjust the size and position of ROI for the tissue section being analyzed (Figure 2-2). Input appropriate experimental parameters such as the step size in X and Y directions (in mm) (Figure 2-3), and name the MSI file in the "Project Name" box (Figure 2-4).
- When drawing an ROI, include a portion of off-tissue that serves as control. Use this off-tissue section for peak picking (Step 5.4).
Note: The desorption diameter (spot size) of the laser on tissue is approximately 150 µm; however, higher spatial resolutions can be obtained by using the oversampling method14,15.
- When drawing an ROI, include a portion of off-tissue that serves as control. Use this off-tissue section for peak picking (Step 5.4).
- Select the appropriate laser frequency from the dropdown box, number of laser pulses per voxel (integer values), and the delay between the laser trigger and mass spectrometer signal acquisition (Figure 2-5). Typically use two laser pulses per voxel at 20 Hz to completely desorb material in an IR-MALDESI MSI experiments, with a 10 msec delay time to allow ions generated to reach the mass analyzer for measurement5.
Note: Choose the highest repetition rate that the laser is capable of operating at. It was recently shown that using higher repetition rate will improve analyte detectability16.
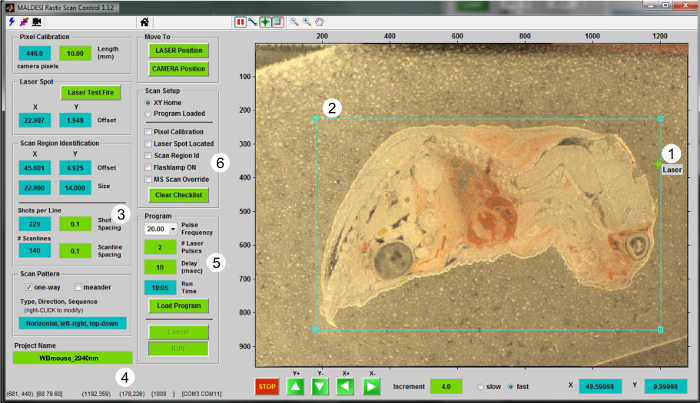
Figure 2. User interface for IR-MALDESI MSI operation. Screenshot of the RASTIR Scan Control program is presented. The steps for performing a MSI experiment are (1) locating the laser spot, (2) drawing an ROI, (3) choosing the stage step size (in mm), (4) giving a name to the file, (5) choosing the correct number of pulses per voxel along with the desired repetition rate, and (6) checking the list for imaging and MS setup. Please click here to view a larger version of this figure.
- Choose mass spectrometer parameters such as ionization mode, electrospray voltage, solvent flow rate, capillary temperature, and the injection time in the mass spectrometer software. Refer to Table 1 for an example of the parameters used in whole-body IR-MALDESI MSI.
Note: Due to the complexity of biological samples and lack of separation methods in MSI analyses, the IR-MALDESI source is coupled to a high resolving power and high mass accuracy instrument. An acquisition method can be loaded for analyses such as parallel reaction monitoring (PRM)7 or polarity switching MSI12. - Once all parameters (laser, stage, and mass spectrometer) have been chosen, place the MS in "handshake" mode for synchronization, and start MS acquisition.
- Using RASTIR imaging software, verify all steps in checklist (Figure 2-6) are completed by checking boxes, and load the program. Once the program has been loaded, the "Run" button will be available. Press Run to start MSI signal acquisition.
- Upon completion of the imaging experiment, stop mass spectrometer acquisition and place the instrument in standby mode. Turn off nitrogen purge to the enclosure, turn off power supply to Peltier plate, and turn off the stage controller and laser.
- Open the access door, remove the electrospray emitter tip from inside the source enclosure, and remove the extended heated metal MS inlet using protective gloves. Caution: The extended metal MS inlet will be very hot.
- Wearing gloves and safety goggles, clean the extended metal capillary inlet by ultrasonic bath in 15% nitric acid for 10 min, then in HPLC-grade water for 10 min, and finally in HPLC-grade methanol for 10 min. Dry the metal capillary inlet under a stream of nitrogen, and reinsert into MS. Note: Dispose solvent in the appropriate waste containers afterward.
| Parameter | Value |
| Ionization Mode | Positive |
| Electrospray Voltage | 3.8-4.2 kV |
| Solvent Flow Rate | 2 μl/min |
| Capillary Temperature | 275 °C |
| Scan Range | m/z 250-1,000 |
| Scan Type | Full Scan |
| Injection Time | 110 msec |
| Resolving Power | 140,000 |
Table 1. Instrument parameters used in whole-body IR-MALDESI MSI.
5. Data Analysis
- Convert the raw data files generated by the instrument to data formats such as mzXML17 or imzML18 using free software such as MSConvert17 and imzML converter19.
- Start MSiReader v1.0, an open-source software developed for analysis of high-resolving power MSI data20, on a dedicated processing computer. Load the imaging file onto MSiReader. For user interface of MSiReader and some of its built-in features, see Figure 3.
- Generate ion maps of analytes of interest by inputting their m/z and choose an appropriate m/z window in parts per million (ppm) or Thomson (Th) (Figure 3-1). For high resolving power (RP) instruments choose a window in the low-ppm range.
Note: The appropriate m/z window depends on the RP and mass measurement accuracy (MMA) of the instrument that IR-MALDESI source is coupled to. - Further interpret the data using the built-in features such as interpolation (Figure 3-2), normalization (Figure 3-3), optical image overlay (Figure 3-4), and peak picking (Figure 3-5)20.
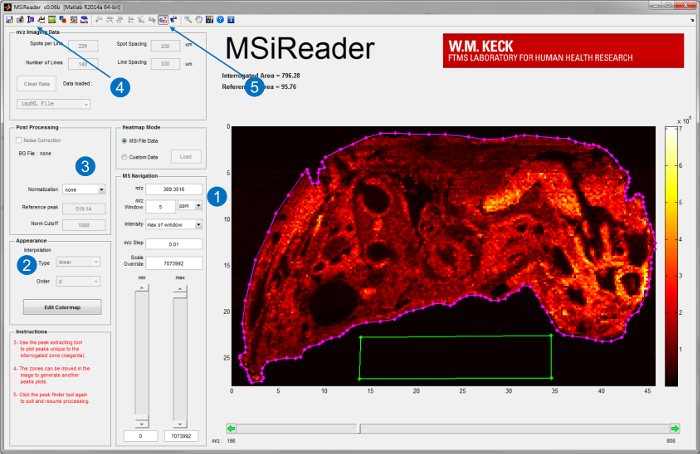
Figure 3. User interface of MSiReader; v1.020. Once a file is loaded into the software, ion maps of analytes of interest are displayed by (1) inputting the m/z and tolerance in ppm or Th. Further analysis such as (2) interpolation or (3) normalization can also be performed. An optical image of the tissue can also be imported and superimposed with the ion maps (4) for better visualization. For untargeted analyses the peak picking function (5) can be used to extract tissue-specific peaks by choosing the area of tissue (magenta line) and a reference area off-tissue (green box). Please click here to view a larger version of this figure.
- Use the peak picking function to generate a list of tissue-specific m/z values using user-defined criteria. Note: The PeakFinder algorithm uses differences in average mass spectrum of two regions. These m/z values can then be searched against metabolite databases such as METLIN21 or LIPID MAPS.22
- Refer to Figure 3 for an example of choosing an interrogated tissue region (magenta polygon ROI) and off-tissue reference region (green polygon ROI).
Wyniki
The images presented in Figure 4 show the spatial distribution of metabolites in different organs in the whole-body tissue section. Unique m/z values to specific regions of the body were found using MSiReader PeakFinder, followed by batch processing for image generation. The image overlay tool (Figure 3-4) was used to align the optical image taken before ice matrix deposition with the resulting ion maps. Cholesterol is observed across all tissue ...
Dyskusje
The protocol above describes the key steps for performing an IR-MALDESI MSI experiment. The matrix application process (Section 3) takes approximately 20 min, which is similar to a typical matrix application process for MALDI MSI experiments by sublimation or spray-coating using a robotic sprayer. Furthermore, IR-MALDESI does not rely on partitioning of analytes into the matrix crystals6, and the ice matrix can be universally used for all analytes regardless of their mass, size, or chemical properties. In addi...
Ujawnienia
The authors declare no competing financial interests.
Podziękowania
The authors thank Professor H. Troy Ghashghaei from NCSU Department of Molecular Biomedical Sciences for providing the whole mouse tissue. The authors also gratefully acknowledge the financial assistance received from National Institutes of Health (R01GM087964), the W.M. Keck foundation, and North Carolina State University.
Materiały
| Name | Company | Catalog Number | Comments |
| IR-MALDESI Source | Custom-made | N/A | Please refer to references 4 and 12 for an in-depth discussion of IR-MALDESI source development. |
| Q Exactive Plus | Thermo Scientific | Q Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer | |
| Water, HPLC Grade | Burdick & Jackson | AH365-4 | |
| Methanol, HPLC Grade | Burdick & Jackson | AH230-4 | |
| Formic Acid | Sigma Aldrich | 56302 | |
| Tunable mid-IR Laser | Opotek Inc. | IR Opolette | Tunable 2,700-3,100 nm IR OPO laser |
| Nitrogen Gas | Arc3 Gases | AG S-NI300-5.0 | Grade 5.0 high purity nitrogen gas cylinder (300) |
| Cryostat | Leica Biosystems | CM 1950 | Cryomicrotome |
| High Profile Microtome Blades | Leica Biosystems | 3802123 | Leica DB80HS |
| Mounting Medium (OCT) | Leica Biosystems | 3801480 | Surgipath FSC 22 mounting medium |
| Cryostat Specimen Disc | Leica Biosystems | 14047740045 | 40 mm diameter |
| Glass Microscope Slides | VWR | 48312-003 | Frosted, selected, pre-cleaned |
Odniesienia
- Mcdonnell, L. A., Heeren, R. M. A. Imaging Mass Spectrometry. Mass Spectrom. Rev. 26, 606-643 (2007).
- Chughtai, K., Heeren, R. M. A. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 110 (5), 3237-3277 (2010).
- Robichaud, G., Barry, J. A., Muddiman, D. C. Atmospheric Pressure Mass Spectrometry Imaging. Encycl. Anal. Chem. , (2014).
- Sampson, J. S., Hawkridge, A. M., Muddiman, D. C. Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 17 (12), 1712-1716 (2006).
- Robichaud, G., Barry, J. A., Garrard, K. P., Muddiman, D. C. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J. Am. Soc. Mass Spectrom. 24 (1), 92-100 (2013).
- Robichaud, G., Barry, J. A., Muddiman, D. C. IR-MALDESI Mass Spectrometry Imaging of Biological Tissue Sections Using Ice as a Matrix. J. Am. Soc. Mass Spectrom. 25 (3), 319-328 (2014).
- Barry, J. A., et al. Mapping Antiretroviral Drugs in Tissue by IR-MALDESI MSI Coupled to the Q Exactive and Comparison with LC-MS/MS SRM Assay. J. Am. Soc. Mass Spectrom. 25 (12), 2038-2047 (2014).
- Rosen, E. P., Bokhart, M. T., Ghashghaei, H. T., Muddiman, D. C. Influence of Desorption Conditions on Analyte Sensitivity and Internal Energy in Discrete Tissue or Whole Body Imaging by IR-MALDESI. J. Am. Soc. Mass Spectrom. 26, 899-910 (2015).
- Nelson, K. A., Daniels, G. J., Fournie, J. W., Hemmer, M. J. Optimization of whole-body zebrafish sectioning methods for mass spectrometry imaging. J. Biomol. Tech. 24 (3), 119-127 (2013).
- Park, J. J., Cunningham, M. G. Thin sectioning of slice preparations for immunohistochemistry. J. Vis. Exp. (3), e194 (2007).
- Bokhart, M. T., Rosen, E., Thompson, C., Sykes, C., Kashuba, A. D. M., Muddiman, D. C. Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using infrared matrix-assisted laser desorption electrospray ionization. Anal. Bioanal. Chem. 407 (8), 2073-2084 (2015).
- Nazari, M., Muddiman, D. C. Polarity Switching Mass Spectrometry Imaging of Healthy and Cancerous Hen Ovarian Tissue Sections by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI). Analyst. 141, 595-605 (2016).
- Hsu, C. C., et al. Design and Application of a Low-Temperature Peltier-Cooling Microscope. J. Pharm. Sci. 85 (1), 70-74 (1996).
- Jurchen, J. C., Rubakhin, S. S., Sweedler, J. V. MALDI-MS imaging of features smaller than the size of the laser beam. J. Am. Soc.Mass Spectrom. 16 (10), 1654-1659 (2005).
- Nazari, M., Muddiman, D. C. Cellular-level mass spectrometry imaging using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) by oversampling. Anal. Bioanal. Chem. 407 (8), 2265-2271 (2015).
- Rosen, E. P., Bokhart, M. T., Nazari, M., Muddiman, D. C. Influence of C-Trap Ion Accumulation Time on the Detectability of Analytes in IR-MALDESI MSI. Anal. Chem. 87, 10483-10490 (2015).
- Kessner, D., Chambers, M., Burke, R., Agus, D., Mallick, P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 24 (21), 2534-2536 (2008).
- Schramm, T., et al. ImzML - A common data format for the flexible exchange and processing of mass spectrometry imaging data. J. Proteomics. 75 (16), 5106-5110 (2012).
- Race, A. M., Styles, I. B., Bunch, J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J. Proteomics. 75 (16), 5111-5112 (2012).
- Robichaud, G., Garrard, K. P., Barry, J. A., Muddiman, D. C. MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J. Am. Soc. Mass Spectrom. 24 (5), 718-721 (2013).
- Smith, C. A., O'Maille, G., et al. METLIN: a metabolite mass spectral database. Ther. Drug. Monit. 27 (6), 747-751 (2005).
- Sud, M., et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 35, D527-D532 (2007).
- Schwartz, S. A., Reyzer, M. L., Caprioli, R. M. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 38 (7), 699-708 (2003).
- Takai, N., Tanaka, Y., Inazawa, K., Saji, H. Quantitative analysis of pharmaceutical drug distribution in multiple organs by imaging mass spectrometry. Rapid Commun. Mass Spectrom. 26 (13), 1549-1556 (2012).
- Liu, J., Gingras, J., Ganley, K. P., Vismeh, R., Teffera, Y., Zhao, Z. Whole-body tissue distribution study of drugs in neonate mice using desorption electrospray ionization mass spectrometry imaging. Rapid Commun. Mass Spectrom. 28 (2), 185-190 (2014).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone