Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
A Protocol for Using Gene Set Enrichment Analysis to Identify the Appropriate Animal Model for Translational Research
W tym Artykule
Podsumowanie
We provide a standardized protocol for the use of gene set enrichment analysis of transcriptomic data to identify an ideal mouse model for translational research.
This protocol can be used with DNA microarray and RNA sequencing data and can further be extended to other omics data if data are available.
Streszczenie
Recent studies that compared transcriptomic datasets of human diseases with datasets from mouse models using traditional gene-to-gene comparison techniques resulted in contradictory conclusions regarding the relevance of animal models for translational research. A major reason for the discrepancies between different gene expression analyses is the arbitrary filtering of differentially expressed genes. Furthermore, the comparison of single genes between different species and platforms often is limited by technical variance, leading to misinterpretation of the con/discordance between data from human and animal models. Thus, standardized approaches for systematic data analysis are needed. To overcome subjective gene filtering and ineffective gene-to-gene comparisons, we recently demonstrated that gene set enrichment analysis (GSEA) has the potential to avoid these problems. Therefore, we developed a standardized protocol for the use of GSEA to distinguish between appropriate and inappropriate animal models for translational research. This protocol is not suitable to predict how to design new model systems a-priori, as it requires existing experimental omics data. However, the protocol describes how to interpret existing data in a standardized manner in order to select the most suitable animal model, thus avoiding unnecessary animal experiments and misleading translational studies.
Wprowadzenie
Animal models are widely used to study human diseases, because of their assumed similarity to humans in terms of genetics, anatomy, and physiology. Moreover, animal models often serve as gatekeepers to clinical therapies and can have a huge impact on the success of translational research. Careful selection of the optimal animal model can reduce the number of misleading animal studies. Recently, the relevance of animal models for translational research has been controversially discussed, particularly because analyzing the same datasets obtained from human inflammatory diseases and related mouse models led to contradictory conclusions 1,2. This discussion revealed a fundamental problem during analyzing omics data: standardized approaches for systematic data analysis are needed in order to reduce biased gene selection and to increase the robustness of interspecies comparisons 3.
Traditionally, the analysis of transcriptomics data (and other omics data) is done at the single-gene level and includes an initial step of gene selection based on stringent cut-off parameters (e.g., fold change >2.0, p value <0.05). However, the setting of initial cut-off parameters often is subjective, arbitrary and not biologically justified, and can even lead to opposite conclusions1,2. Furthermore, initial gene selection generally restricts the analysis to a few highly up- and downregulated genes and is thus not sensitive enough to include the majority of genes that were differentially expressed to a lesser extent.
With the rise of the genomics era in the early 2000s and the increasing knowledge of biological pathways and contexts, alternative statistical approaches were developed that allowed to circumvent the limitations of single-gene level analyses. Gene set enrichment analysis (GSEA)4, which is one of the widely accepted methods for the analysis of transcriptomics data, makes use of a-priori defined groups of genes (e.g., signaling pathways, proximal location on a chromosome etc.). GSEA first maps all detected unfiltered genes to the intended gene sets (e.g., pathways), irrespective of their individual change in expression. This approach thus also includes moderately regulated genes that would otherwise be lost with single-gene level analyses. The additive change in expression within gene sets is subsequently performed using running sum statistics.
Despite its wide use in medical research, GSEA and related set enrichment approaches are not self-evidently taken into account for the analysis of complex omics data. Here, we describe a protocol for comparing omics data from human samples with those from mouse models in order to identify the ideal model for translational studies. We demonstrate the applicability of the protocol based on a collection of mouse models that are used for mimicking human inflammatory disorders. However, this analysis pipeline is not restricted to human-mouse comparisons and is amendable to further research questions.
Protokół
1. Download of the GSEA Software and the Molecular Signatures Database
- Go to the official GSEA Broad Institute website (http://software.broadinstitute.org/gsea/index.jsp) and register to get access to the GSEA software tool and the Molecular Signatures Database (MSigDB).
- Download the javaGSEA desktop application or an alternative software option (e.g., R script).
NOTE: All options implement exactly the same algorithm. The GSEA software is freely available to individuals in academia and industry for internal research purposes. - For further details on the GSEA software go to the documentation website (http://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/Main_Page) and the GSEA user guide (http://software.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html).
- Download the Molecular Signatures Database (MSigDB) from the GSEA website to get access to individual gene set collections.
NOTE: The MSigDB is a collection of annotated gene sets for use with the GSEA software or other purposes. Gene sets can be divided according to signaling pathways, gene ontology terms, cis-regulatory motifs, experimental signatures and others. Genes from the MSigDB are always named by their official HUGO (Human Genome Organisation) gene symbol. For the comparison of pathway regulation between a given human disorder and different mouse models it is recommended to download the 'all canonical pathways, gene symbols' file (c2.cp.v5.2.symbols.gmt). This file comprises gene sets that were annotated and organized into signaling pathways by KEGG 5,6, Reactome 7,8 and BioCarta 9. The string 'v5.2' represents the version information of the collection. Make sure to download the latest version of the files. The MSigDB is freely available to individuals in academia and industry for internal research purposes. It is not needed to download the MSigDB, if internet connection is provided during the analysis. In this case the MSigDB can directly be chosen within the GSEA user interface. - Download DNA chip (array) annotations files from the GSEA website to translate array-specific probe identifiers to general HUGO gene symbols (e.g., Mouse430_2.chip).
NOTE: It is not needed to download the DNA chip annotations, if internet connection is provided during the analysis. In this case the DNA chip annotations can directly be chosen within the GSEA user interface. The protocol can also be used with RNA sequencing data. In this case, it is not needed to download annotation files. Instead, use the GSEA preranked tool for analyzing the gene expression data (see step 4.12).
2. Download Experimental Gene Expression Data for the Human Disorder and Appropriate Animal Models
- Identify experimental gene expression (transcriptomics) studies for the human disorder of choice (e.g., gene expression profiles of leukocytes derived from patients with septic disorder, GSE9960).
- Likewise, search for several animal models that are supposed to be compared with the human studies (e.g., gene expression profiles of blood cells derived from mice after injection of Staphylococcus aureus (S. aureus), GSE20524). At this step use the prior knowledge for the preselection of animal models that might be suitable for mimicking the human situation.
- For this purpose refer to literature and databases such as the Gene Expression Omnibus (GEO) database 10 or ArrayExpress 11 and download the normalized transcriptomics data of interest. Save the data as text files on the local hard disk. For the GEO database, download of tab-delimited series matrix text files is recommended. Also take note of the platform (array type) used for that study, since this information is needed for translating the array-specific probe identifiers to general HUGO gene symbols.
NOTE: Ensure enough memory for data storage, as transcriptomics data sets usually comprise several hundred MB.
3. Data Handling and Formatting
- Before importing experimental gene expression data into the GSEA software tool, consider the required data structure. For each study manually create two different files: 1) a gene expression data file containing measurement values for various genes and samples, and 2) a phenotype file containing sample labels to group individual samples (e.g., to treatment groups).
For further details and data structure options go to the GSEA data format page (http://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/Data_formats).
NOTE: Generally, all forms of transcriptomics data are compatible with the protocol, including DNA microarray experiments, RNA-seq or ChIP-seq studies. In case of using DNA microarray experiments, the gene expression data file should contain array-specific probe identifier or HUGO gene symbols for each gene (probe identifiers will be translated to HUGO gene symbols during analysis, see steps 1.5 and 4.10). In case of using RNA-seq or ChIP-seq data, manually calculated group metrics for gene expression data (e.g., group mean ratio) should be used instead of individual sample data. These group metrics should then be analyzed with the GSEA preranked tool (see step 4.12). Gene expression data have to be normalized as usual before importing into the GSEA software. The type of normalization (e.g., quartile or cubic spline) is generally left to the researcher. - Gene expression data file: Use the tab-delimited text file (*.txt) format for describing an expression dataset as depicted in Figure 1A. See also the supported example file GSE20524_expression.txt.
NOTE: The gene expression data file contains expression values for all detectable genes (or probes), also for genes that might not be differentially expressed. The file therefore typically comprises many thousands of genes. It is organized as depicted in Figure 1A. The first line contains the label name (e.g., gene symbol or probe ID) followed by the identifier for each sample in the dataset (e.g., sample 1, sample 2 etc.). The remainder of the file contains expression values for each of the genes and for each sample in the dataset. The GSEA software tool performs calculations for group metrics (e.g., group mean ratio or signal-to-noise-ratio), therefore it is recommended to include data for each individual sample. Alternatively, it is possible to use externally calculated group metrics for gene expression data (see Figure 1B). - Phenotype file: Create a separate file for defining and labeling groups that comprise individual samples as depicted in Figure 2. Use spaces or tabs to separate the fields. Save it in a CLS (C++ Class Definition) file format. See also the supported example file GSE20524_pheno_infection.cls.
NOTE: The first line contains the total number of samples and further the number of groups (Figure 2). While the number of samples should correspond to the gene expression data file (see 3.2), the number of groups depends on the study design. The third field of the first line is always '1'.
The second line in a CLS file contains the name for each group. The line should begin with a pound sign (#) followed by a space (Figure 2).
The third line contains a group label for each sample. The group label can be an arbitrary number or text. It is only the order of the labels that determines the association of each sample to the groups: The first label used is assigned to the first group on the second line; the second unique label is assigned to the second group and so on. Ensure that each sample of the same group has the same label at this step, and that the number of labels is the same as the number of samples specified in the first line. Finally, save the file as tab-delimited text file (*.txt) and manually change the file name extension to (*.cls). - (optional) Gene Set Database files: Define custom gene sets. Use the tab-delimited GMT (Gene Matrix Transposed) file format for gene sets as depicted in Figure 3. Also see the supported example file Gene_sets_Inflammation_BIOCARTA_KEGG_REACTOME.gmt.
NOTE: Defining custom gene sets can be useful for instance to restrict the gene set enrichment analysis to pathways of special interest (e.g., immunology signaling for sepsis studies), or for de novo defining own gene sets (e.g., activated and inhibited genes in studies that have to be compared). The file is organized as depicted in Figure 3. In the GMT format, each row represents a gene set (Figure 3). Each gene set is described by a name, a description, and the genes in the gene set. The first column contains unique gene set names. The second line may optionally contain a description of the gene set. The following columns contain the gene names (official HUGO gene symbols) of the corresponding gene set. Finally, save the file as tab delimited text file (*.txt) and manually change the file name extension to (*.gmt).
4. Performing the GSEA
- Open the GSEA software tool (see 1.2).
- Click the 'load data' button on the left side of the main window (Figure 4A). A new tab will open for importing the required data files (Figure 4B). Browse in the new tab to the gene expression data (*.txt) file (see 3.2), the phenotype (*.cls) file (see 3.3) and, optionally, to the custom gene sets (*.gmt) file (Figure 4B).
- In case GSEA cannot connect to the internet, also load the downloaded MSigDB (*.gmt) files (e.g., c2.cp.v5.2.symbols.gmt for pathways, see 1.4) and the DNA chip (array) annotations (*.chip) files (e.g., Mouse430_2.chip, see 1.5). Successfully imported data appear in the 'load data' section (Figure 4C).
NOTE: Each gene expression study must be analyzed with GSEA individually. The comparison between two studies (e.g. human disorder vs. mouse model) will be performed at step 5.
- In case GSEA cannot connect to the internet, also load the downloaded MSigDB (*.gmt) files (e.g., c2.cp.v5.2.symbols.gmt for pathways, see 1.4) and the DNA chip (array) annotations (*.chip) files (e.g., Mouse430_2.chip, see 1.5). Successfully imported data appear in the 'load data' section (Figure 4C).
- Click the 'Run GSEA' button on the left side of the main window. A new tab will open in order to set the parameters for the analysis (Figure 4D). The tab is subdivided into three parts: required fields, basic fields and advanced fields.
- In the required fields, first choose the expression dataset loaded in step 4.2 (Figure 4D).
- Choose the gene sets database, either from the connected website or from the manually imported gene set file (Figure 4D).
- Edit the phenotype labels to select the groups of samples that are supposed to be compared to each other (e.g., S. aureus treatment vs. healthy control) (Figure 4D).
- Collapse dataset to gene symbols (=true) in order to translate the probe identifiers in the expression dataset to official HUGO gene symbols used in the gene sets database. Select false, if the expression dataset already contains HUGO gene symbols (Figure 4D).
- Set the number of permutations to default setting at 1,000 (Figure 4D).
NOTE: For higher numbers the computing time will increase considerably. - Change the permutation type to 'gene set', since phenotype permutation is only recommended when there are more than seven samples in every phenotype (Figure 4D).
- Finally, select the chip platform used for generating the gene expression data, either from the connected website or from the manually imported DNA chip (array) annotations file (Figure 4D).
NOTE: This step is only necessary, if probe identifiers are used in the uploaded expression dataset. - In the basic fields edit at least the analysis name and the save results in this folder section to find again the results file (Figure 4D). In addition, further statistical parameters can be changed. For further details on the parameters and the advanced fields section please go to the GSEA user guide (http://software.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html).
- (Optional): In the case externally calculated group metrics for gene expression data (e.g., group mean ratio) have to be used instead of individual sample data, use the GSEA preranked tool. The analysis will then be conducted based on a simple list of genes assigned with pre-calculated group metrics that are used to rank the genes. After loading the alternative gene expression file go to the main navigation bar and click on Tools/GseaPreranked. Similarly, a new tab will open for setting the parameters for the analysis (Figure 4E).
NOTE: Using the GSEA preranked tool is recommended for studies that do not have individual sample-specific gene expression data. That could be the case if special statistics or normalization procedures were performed on the data leading to group mean values instead of individual sample data. Using the GSEA preranked tool is recommended for RNA sequencing data. Normalize the RNA sequencing expression data and calculate group metrics for the samples (e.g., log of fold change), that can be used to rank the genes according their expression. - Click the 'Run' button on the right bottom of the window.
NOTE: The analysis then may take up to several minutes depending on the computing speed. Follow the progress of the analysis in the GSEA reports section on the left bottom of the window. After finishing the analysis, the status 'success' appears in the GSEA reports section. - Click on the succeeded analysis in the GSEA reports section to open the analysis results.
NOTE: A new navigation menu will open in a browser window that summarizes all results and parameter settings (Figure 5). The upper two sections of the navigation menu comprise gene set enrichment results for the defined groups (e.g., enrichment in S. aureus treated samples or healthy control samples). The first lines of both sections show a summary of the statistic results. Gene sets that are significantly enriched at a false-discovery rate (FDR) below 25% are regarded as enriched in the following interpretation. Further details on the interpretation of the analysis can be found in the GSEA user guide (http://software.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html). - Click on the detailed enrichment results in excel format to export the analysis results to a spreadsheet (Figure 6A). Export the detailed enrichment results in excel separately for both phenotypes (Figure 5) and join the results data in one spreadsheet file. For subsequent comparison between gene expression data of several studies, maintain at least the name of the gene set (column A), its normalized enrichment score (NES) (column F) and its FDR (false discovery rate) value (column H) (Figure 6B).
NOTE: The spreadsheet file contains huge data for each of the analyzed gene set, including the name of the gene set (column A), its size (that is, the number of genes detected in the gene expression data, column D), its NES (a quantitative measure of the direction and extent of the enrichment, column F), its nominal p value (uncorrected, column G) and its FDR value (corrected for multiple hypothesis testing, column H). For further details on the interpretation please refer to the GSEA user guide (http://software.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html). - Repeat the gene set enrichment analysis (steps 4.1 to 4.15) for the second study (e.g., S. aureus GSE9960) and for all further studies that are supposed to be compared to each other. Include as many human clinical studies and different mouse models as possible to identify the optimal mouse model for the translational research question.
5. Comparing the GSEA Results
- To identify the optimal animal model for mimicking the human situation compare the GSEA results of all studies to each other. Use the enrichment scores and the FDR values to classify the pathways (gene sets) as activated (NES >0, FDR <25%), inhibited (NES <0, FDR <25%) or none of both (FDR >25%). For each comparison of two studies, count the number of realizations of the nine possible combinations of pathway regulation as indicated by a 3x3 contingency table (Figure 7A).
- Assess the correlation between two studies by calculation of the positive predictive value (ppv) and the negative predictive value (npv), which is by definition the part of pathways that show the same regulation (activated or inhibited) in two studies.
- Calculate ppv and npv according to the following formulas (1) and (2):
(1)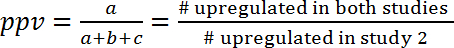
(2)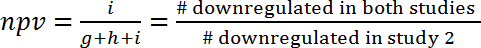
NOTE: Since the overlap could be purely coincidental, the ppv and the npv have to be further compared to the values expected by chance. This approach allows the estimation of the amount of information that can be gained from one study for predicting the effects in another study. For instance, if the regulation processes in two models were independent from one another (and only overlap by chance), and if in the first model 10% of the pathways were upregulated, than the ppv to the second model would also be 10% and there was no additional gain of information. On the other side, if both models were linked by common regulation mechanisms, then the ppv (and npv) would be significantly greater than expected by chance. For example, for the prediction of gene expression changes during human sepsis (GSE9960) from effects in a murine S. aureus injection model (GSE20524), the ppv is 43% (6/(6+8+0)) and the npv is 61% (11/(0+7+11)). In other words, 43% of the activated pathways in the murine S. aureus injection model (GSE20524) are also activated during human sepsis (GSE9960). Similarly, 61% of the inhibited pathways in the murine S. aureus injection model (GSE20524) are also inhibited during human sepsis (GSE9960) (Figure 7B). ppv and npv can also be determined for the inverse constellation (that means predicting from study 1 to study 2).
- Calculate ppv and npv according to the following formulas (1) and (2):
- To calculate the overlap by chance refer to the 3x3 contingency table (Figure 7) and calculate ppvchance and npvchance according to the following formulas (3) and (4):
(3)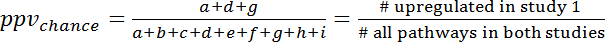
(4)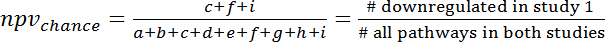
NOTE: For example, for the prediction of gene expression changes during human sepsis (GSE9960) from effects in a murine S. aureus injection model (GSE20524) the ppvchance is 13% (8/64) and the npvchance is and 22% (14/64). - Calculate the gain of ppv vs. chance by subtracting ppvchance from ppv. Calculate accordingly for the npv:
(5)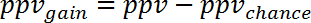
(6)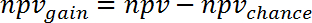
NOTE: For example, for the prediction of gene expression changes during human sepsis (GSE9960) from effects in a murine S. aureus injection model (GSE20524) the change in ppv and npv vs. chance is +30% (43% - 13%) and +39% (61% - 22%), respectively. - Calculate the gain of information that can be obtained from study 2 regarding study 1 by averaging ppvgain and npvgain:
(7)
- Use the contingency table defined in step 5.1 of a pair of studies (study1.pathway, study2.pathway) to calculate the p value by a chi-squared test.
Store the data of the contingency table in a matrix X. Perform the chi-squared test, e.g., by use of the R function chisq.test.
NOTE: For example, comparing the selected human sepsis study (GSE9960) with a murine S. aureus injection model (GSE20524) shows a statistically significant overlap in inflammatory pathway regulation:
> chisq.test(X,simulate.p.value=F)$p.value
3.82e-07
6. Identifying the Optimal Animal Model
- Compare the GSEA results for all combinations of the studies that were selected for the analysis.
NOTE: It is also recommended to compare the (similar) human studies to one another as well as the different animal studies to one another. This comparison can provide insight into the intraspecies variance of the clinical studies (or disorders) and the different animal models. It is expected that the clinical studies should show an acceptable overlap and a significant information gain, because otherwise the clinical studies might be too heterogeneous to find an animal model that can mimic the human situation. In this case, it is recommended to include only human studies that are similar to each other for the identification of suitable animal models. - Sort all combinations by the gain of information (step 5.5). For the comparison of many datasets, use a matrix and visualize the findings by use of a colored heatmap or the like (Figure 8).
- Select the animal model with the highest gain of information. In order to assess the significance of the gain of information, also take the chi-squared test (step 5.6) into account.
NOTE: Animal models should only be regarded as appropriate if the gain of information is substantial and if the p value of the chi-squared test is below the significance level. User-defined thresholds will generally depend on several factors: 1) the pre-study knowledge on the transferability of the results from animal model to humans (e.g. similar physiology), 2) the expected benefit for humans by a presumed success, 3) the practical applicability of that animal experiment, and 4) the expected pain, suffering, or harm inflicted on the laboratory animals.
Wyniki
The GSEA workflow and screenshots of exemplary data are demonstrated. Figure 1 shows the gene expression data file that contains the transcriptomic data of interest. For every study a descriptive phenotype file is required that is shown in Figure 2. Annotated gene sets (e.g., pathways) are defined in the gene set database file (Figure 3). Figure 4 shows a step-b...
Dyskusje
Animal models have long been applied for the investigation of disease mechanisms and the development of novel therapeutic strategies. However, skepticism regarding the predictivity of animal models started to spread following failures of clinical trials12. Furthermore, controversial discussions about appropriate strategies for analyzing and interpreting big omics data from preclinical trials were raised by opposite conclusions drawn from the same data after applying differing data analysis strateg...
Ujawnienia
The authors declare that they have no competing financial interests.
Podziękowania
This work was financed by the German Federal Institute for Risk Assessment (BfR).
Materiały
| Name | Company | Catalog Number | Comments |
| Excel | Microsoft Corporation |
Odniesienia
- Seok, J., et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 110 (9), 3507-3512 (2013).
- Takao, K., Miyakawa, T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 112 (4), 1167-1172 (2015).
- Weidner, C., Steinfath, M., Opitz, E., Oelgeschläger, M., Schönfelder, G. Defining the optimal animal model for translational research using gene set enrichment analysis. EMBO Mol Med. 8 (8), 831-838 (2016).
- Subramanian, A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102 (43), 15545-15550 (2005).
- Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44 (D1), D457-D462 (2016).
- Kanehisa, M., Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27-30 (2000).
- Fabregat, A., et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 44 (D1), D481-D487 (2016).
- Croft, D., et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 42 (Database issue), D472-D477 (2014).
- Nishimura, D. BioCarta. Biotech Software & Internet Report. 2 (3), 117-120 (2001).
- Edgar, R., Domrachev, M., Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 (1), 207-210 (2002).
- Kolesnikov, N., et al. ArrayExpress update--simplifying data submissions. Nucleic Acids Res. 43 (Database issue), D1113-D1116 (2015).
- Cohen, J., et al. Sepsis: a roadmap for future research. Lancet Infect Dis. 15 (5), 581-614 (2015).
- Spinelli, L., Carpentier, S., Montanana Sanchis, F., Dalod, M., Vu Manh, T. P. BubbleGUM: automatic extraction of phenotype molecular signatures and comprehensive visualization of multiple Gene Set Enrichment Analyses. BMC Genomics. 16 (1), 814 (2015).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone