Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Genetic Analysis of Hereditary Transthyretin Ala97Ser Related Amyloidosis
W tym Artykule
Podsumowanie
Here, we present a protocol to confirm the presence of point mutation for the diagnosis of hereditary transthyretin amyloidosis, using Ala97Ser, the most common endemic mutation in Taiwan, as an example.
Streszczenie
Genetic testing is the most reliable test for hereditary transthyretin related amyloidosis and should be performed in most cases of transthyretin amyloidosis (ATTR). ATTR is a rare but fatal disease with heterogeneous phenotypes; therefore, the diagnosis is sometimes delayed. With increasing attention and broader recognition on early manifestations of ATTR as well as emerging treatments, appropriate diagnostic studies, including the transthyretin (TTR) genetic test, to confirm the types and variants of ATTR are therefore fundamental to improve the prognosis. Genetic analyses with polymerase chain reaction (PCR) methods confirm the presence of TTR point mutations much more quickly and safer than conventional methods such as southern blot. Herein, we demonstrate genetic confirmation of the ATTR Ala97Ser mutation, the most common endemic mutation in Taiwan. The protocol comprises four main steps: collecting whole blood specimen, DNA extraction, genetic analysis of all four TTR exons with PCR, and DNA sequencing.
Wprowadzenie
Transthyretin (TTR) amyloidosis (ATTR) is the most common form of hereditary systemic amyloidosis1, and can be caused by an autosomal dominantly inherited mutation in the transthyretin (TTR) gene2. TTR mutations destabilize the tetrameric protein structure and lead to its dissociation into monomers that reassembles into amyloid fibrils2. More than 100 amyloidogenic TTR mutations have been reported worldwide1. Genetic analyses with polymerase chain reaction (PCR) methods confirm the presence of TTR point mutation and have advantages including avoiding the handling of radioactively labeled probes in comparison with southern blot3. PCR is a fast, easy, cheap, and reliable technique that has been applied to numerous fields in modern sciences4.
The early diagnosis of this progressive and fatal disease is challenging given its phenotypic heterogeneity. With increasing attention and broader recognition on the early manifestations of ATTR as well as the emerging treatments5, appropriate diagnostic studies including TTR genetic test are therefore critically fundamental to improve prognosis. Furthermore, different mutations are associated with different penetrance of the trait, age of onset, patterns of progression, disease severity, median survival, efficacy of liver transplantation, or TTR stabilizers2,6, and variable degrees of neurological and cardiological involvement, which have great implications for genetic counseling7,8,9. Besides, a highly accurate genetic test is the only tool that differentiates the two distinct types of ATTR: hereditary (mutant) and wild type (non-mutant form, senile systemic amyloidosis, SSA)7. It is imperative to confirm the types of ATTR, because the therapies vary widely2. Therefore, there is an increasing necessity to describe the stepwise protocol of the TTR genetic test.
The molecular approach to detect the mutation will be illustrated using Ala97Ser, the most common endemic mutation in Taiwan, as an example. Modifications in the DNA extraction step reduce the amount of the three solutions used and yields a sufficient amount of DNA. In this protocol, all four TTR exons were analyzed, while regions including 5' upstream, 3' downstream, promoters, introns, and untranslated regions (UTR) were not sequenced.
Protokół
The testing performed in the laboratory was carried out in accordance with the requirements of the Clinical Laboratory Improvement Amendments (CLIA) of 1988, the regulations approved by the Institutional Review Board of Chang Gung Memorial Hospital and University (License no. 100-4470A3 and 104-2462A3). Informed consent was obtained from all patients.
1. Blood Specimen Collection
- Collect whole blood into commercially available EDTA-treated tubes. Mix gently and store blood sample at 4 °C until processing.
2. DNA Extraction from Peripheral Blood
Use a DNA Extraction Kit for the genomic DNA extraction (see Table of Materials).
- Add 10 mL of Solution 1 to the 4.0 mL blood sample. Incubate the sample on ice for 10 min.
- Centrifuge the sample at 2,000 x g for 5 min at 4 °C. Remove and discard supernatant carefully. Resuspend the pellet in 3 mL of Solution 1.
- Centrifuge the sample at 2,000 x g for 5 min at 4 °C again and discard supernatant. Resuspend the pellet in 3 mL of Solution 2.
- Add 10 µL of pronase stock solution (225 mg/mL) to get a final concentration of 100 µg/mL and mix well. Incubate at 37 °C overnight.
- Chill the tube on ice for 10 min. Add 0.8 mL of Solution 3 to the sample and invert 3 - 5 times. Place sample on ice for 5 min.
- Centrifuge the sample at 3,000 x g for 15 min at 4 °C. Carefully transfer the supernatant to a 15 mL sterile conical centrifuge tube.
- Add 6 µL of RNase stock solution (10 mg/mL) to get a final concentration of 20 µg/mL. Incubate at 37 °C for 15 min.
- Add 2.5 mL of isopropyl alcohol and mix thoroughly by gently inverting several times to precipitate DNA (strands of white, flocculent material will form). Using a large-bore pipet, transfer the DNA precipitant to a new 1.5 mL centrifuge tube.
- Centrifuge at 12,000 x g for 3 min at 4 °C. Discard supernatant carefully. Wash DNA pellet by adding 500 µL of 70% ethanol.
- Centrifuge at 12,000 x g for 1 min and discard supernatant. Air dry DNA pellet at room temperature. Add 100 µL of ddH2O and incubate at 65 °C for 15 min to resuspend DNA.
3. Genomic DNA Quantification
Use a spectrophotometer (see Table of Materials) to detect the quantity and quality of the genomic DNA.
- Log into the computer attached to the spectrophotometer machine. Open the software.
- Initialize the spectrophotometer machine:
- Click "Nucleic Acid" on the spectrophotometer software.
- Clean the pedestal with a disposable paper wipe and purified water.
- Blank the spectrophotometer:
- Load 2 µL of purified water on the pedestal.
- Lower the upper arm of the spectrophotometer and click "Blank" to calibrate it.
- When it is done, lift the upper arm and dry the pedestal with a wipe.
- Measure the sample:
- Click "DNA-50" on Sample Type.
- Load 2 µL of DNA sample on the pedestal.
- Lower the upper arm and click "Measure" to measure the A260/A280 ratio and the concentration of the sample.
- Clean the pedestal in between each run with a wipe and purified water.
- Click "Print Screen" to collect the data.
4. Genetic analyses of mutations
Amplify the target DNA with the PCR4. Perform PCR in a 25 µL reaction mixture using a DNA Polymerase kit (see Table of Materials). Table 1 shows the TTR gene intronic primers.
| gene | Primer sequence |
| Exon1 | F: 5’-TCAGATTGGCAGGGATAAGC-3’ R: 5’-GCAAAGCTGGAAGGAGTCAC-3’ |
| Exon2 | F: 5’-TCTTGTTTCGCTCCAGATTTC-3’ R: 5’-TCTACCAAGTGAGGGGCAAA-3’ |
| Exon3 | F: 5’-GTGTTAGTTGGTGGGGGTGT-3’ R: 5’-TGAGTAAAACTGTGCATTTCCTG-3’ |
| Exon4 | F: 5’-GACTTCCGGTGGTCAGTCAT-3’ R: 5’-GCGTTCTGCCCAGATACTTT-3’ |
Table 1. TTR gene intronic primers. (NCBI Reference Sequence: NC_000018.10)
- Add the reagents in Table 2 to a 0.2 mL PCR tube.
| Components | Volume (µL) | Final concentration |
| 10x Buffer | 2.5 | 1x |
| MgCl2 (25 mM) | 1.5 | 1.5 mM |
| 360 GC Enhancer | 1 | - |
| 360 DNA Polymerase | 0.2 | 2 U/reaction |
| dNTP Mix (25 mM each) | 2 | 2 mM each |
| Primer F (10 µM) | 1 | 0.4 µM |
| Primer R (10 µM) | 1 | 0.4 µM |
| DNA (100 ng) | 2 | |
| PCR-grade water | 13.8 | - |
| Total Volume | 25 | - |
Table 2. PCR reaction conditions.
- Mix gently and briefly centrifuge on a benchtop centrifuge to collect all components to the bottom of the tube. Place tube in a thermocycler.
- Perform PCR for 30 cycles. Each cycle consists of a denaturation step for 30 s at 94 °C, primer annealing for 30 s at 58 °C, and polymerase extension for 45 s at 72 °C (see Table 3).
| Step | Temperature (°C) | Time | cycle |
| Initial denaturation | 95 | 5 min | 1 |
| DNA denaturation step | 94 | 30 s | 30 cycles |
| Primer annealing step | 58 | 30 s | 30 cycles |
| Polymerase extension step | 72 | 45 s | 30 cycles |
| Post elongation step | 72 | 10 min | 1 |
| End of PCR cycling | 4 | Indefinite |
Table 3. Cycling conditions for amplifying PCR products.
- Validate the PCR product by visualizing with gel electrophoresis:
- Prepare a 1.2% agarose gel:
- Mix 1.2 g of agarose powder with 100 mL of 0.5x TBE in a microwavable flask. Microwave for 2 min until the agarose is completely dissolved. Cool down the agarose mixture to 55 °C.
- Add 2 µL of a ethidium bromide solution (10 mg/mL) to the agarose mixture and mix gently. Pour the agarose mixture into a gel tray to about 5 - 7 mm, then add the comb(s) in place. Allow it to solidify (at least 30 min) and carefully remove comb(s).
- Load samples and run an agarose gel:
- Place gel and tray into the electrophoresis cell. Fill the electrophoresis cell with 0.5x TBE until the gel is covered.
- Carefully load 2 µL of a 100 bp DNA ladder (see Table of Materials) into the one lane of the gel as molecular size marker. Load dye/sample mixture (5 µL of PCR product mix with 1 µL 6x loading dye) into the additional lanes.
- Turn on the power supply. Run the gel for 25 min at 100 V.
- Remove the gel from the electrophoresis cell. Visualize the DNA fragments under UV light in an imaging system.
- Prepare a 1.2% agarose gel:
5. DNA sequencing
- Purify PCR products using a commercial kit and sequence commercially or with an automatic sequencer (see Table of Materials).
- Submit nucleotide sequences to the NCBI BLAST server to compare the nucleotide query to the nucleotide databases10. After the results are displayed, perform sequence alignments and identify the expected mutation.
Wyniki
Agarose gel electrophoresis of two patients and one healthy individual revealed bands of the expected sizes, including a 454 bp PCR product for exon 4 of the TTR gene (Figure 1).
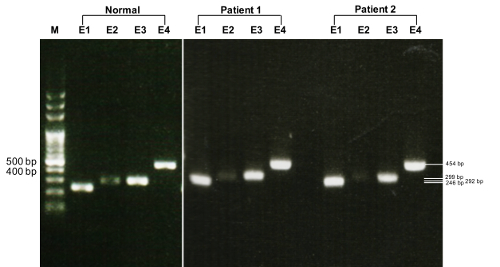
Figure 1. Gel electrophoresis depicting PCR amplified TTR gene. Normal: a healthy indiv...
Dyskusje
There are two critical steps within the protocol. First, in order to have sufficient number of white blood cells, a hemodiluted specimen should be avoided11. Second, the use of appropriate PCR primers is fundamental to obtain reliable results12. We used the Primer-BLAST web tool to design the primers4,13; a minimum of 40 base pairs on each side of the four TTR exons should be covered. We also run BLAST on NCBI to ch...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
We wish to thank Miss Shin-Fun Wu for her help in the experiments. This study was supported by a grant from the Chang Gung Medical Research Program (CMRPG3C0371, CMRPG3C0372, CMRPG3C0373) and IRB 100-4470A3, 104-2462A3, Taiwan.
Materiały
| Name | Company | Catalog Number | Comments |
| EDTA-treated tubes | BD | ||
| DNA Extraction Kit | Stratagen | 200600 | |
| NanoDrop ND2000 spectrophotometer | Thermo Fisher Scientific | NanoDrop 2000 | |
| Delicate Task Wipers | Kimberly-Clark | Kimtech Science Kimwipes | |
| AmpliTaq Gold 360 DNA Polymerase kit | Applied Biosystems | 4398823 | |
| TTR gene intronic primers | Exon1 | F: 5’-TCAGATTGGCAGGGATAAGC-3’ | |
| Exon1 | R: 5’-GCAAAGCTGGAAGGAGTCAC-3’ | ||
| Exon2 | F: 5’-TCTTGTTTCGCTCCAGATTTC-3’ | ||
| Exon2 | R: 5’-TCTACCAAGTGAGGGGCAAA-3’ | ||
| Exon3 | F: 5’-GTGTTAGTTGGTGGGGGTGT-3’ | ||
| Exon3 | R: 5’-TGAGTAAAACTGTGCATTTCCTG-3’ | ||
| Exon4 | F: 5’-GACTTCCGGTGGTCAGTCAT-3’ | ||
| Exon4 | R: 5’-GCGTTCTGCCCAGATACTTT-3’ | ||
| thermocycler | Applied Biosystems | GeneAmp PCR System 9700 | |
| electrophoresis cell | ADVANCE | Mupid-2plus | |
| DNA ladder | Protech | PT-M1-100 | |
| dye | BioLabs | B7021 | |
| AlphaImager EC | Alpha Innotech | AlphaImager EC | |
| automatic sequencer | Applied Biosystems | 3730xl DNA Analyzer |
Odniesienia
- Planté-Bordeneuve, V., Said, G. Familial amyloid polyneuropathy. The Lancet Neurology. 10 (12), 1086-1097 (2011).
- Planté-Bordeneuve, V., et al. Long-term treatment of transthyretin familial amyloid polyneuropathy with tafamidis: a clinical and neurophysiological study. Journal of Neurology. 264 (2), 268-276 (2017).
- Nichols, W. C., Benson, M. D. Hereditary amyloidosis: detection of variant prealbumin genes by restriction enzyme analysis of amplified genomic DNA sequences. Clinical Genetics. 37 (1), 44-53 (1990).
- Lorenz, T. C. Polymerase Chain Reaction: Basic Protocol Plus Troubleshooting and Optimization Strategies. Journal of Visualized Experiments. (63), e3998 (2012).
- Hsu, H. C., et al. Phenotypic expressions of hereditary Transthyretin Ala97Ser related Amyloidosis (ATTR) in Taiwanese. BMC Neurology. 17 (1), 178 (2017).
- Barroso, F. A., et al. Long-term safety and efficacy of tafamidis for the treatment of hereditary transthyretin amyloid polyneuropathy: results up to 6 years. Amyloid. 24 (3), 194-204 (2017).
- Rapezzi, C., et al. Disease profile and differential diagnosis of hereditary transthyretin-related amyloidosis with exclusively cardiac phenotype: an Italian perspective. European Heart Journal. 34 (7), 520-528 (2013).
- Mariani, L. L., et al. Genotype-phenotype correlation and course of transthyretin familial amyloid polyneuropathies in France. Annals of Neurology. 78 (6), 901-916 (2015).
- Hammarström, P., Jiang, X., Hurshman, A. R., Powers, E. T., Kelly, J. W. Sequence-dependent denaturation energetics: A major determinant in amyloid disease diversity. Proceedings of the National Academy of Sciences of the United States of America. 99, 16427-16432 (2002).
- Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., Madden, T. L. NCBI BLAST: a better web interface. Nucleic Acids Research. 36, 5-9 (2008).
- Fox, A. J., et al. Next generation sequencing for the detection of actionable mutations in solid and liquid tumors. Journal of Visualized Experiments. (115), (2016).
- Date, Y., Nakazato, M., Kangawa, K., Shirieda, K., Fujimoto, T., Matsukura, S. Detection of three transthyretin gene mutations in familial amyloidotic polyneuropathy by analysis of DNA extracted from formalin-fixed and paraffin-embedded tissues. Journal of the Neurological Sciences. 150 (2), 143-148 (1997).
- Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., Madden, T. L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 13, 134 (2012).
- Green, M. R., Sambrook, J. . Molecular cloning: A laboratory manual. , (2012).
- Buckingham, L., Flaws, M. L. . Molecular diagnostics: Fundamentals, methods, and clinical applications. , (2012).
- Tan, S. C., Yiap, B. C. DNA, RNA, and Protein Extraction: The Past and The Present. Journal of Biomedicine and Biotechnology. 2009, (2009).
- Rapezzi, C., et al. Systemic cardiac amyloidoses: disease profiles and clinical courses of the 3 main types. Circulation. 120 (13), 1203-1212 (2009).
- Gertz, M. A., Dispenzieri, A., Sher, T. Pathophysiology and treatment of cardiac amyloidosis. Nature Reviews Cardiology. 12 (2), 91-102 (2015).
- Vrana, J. A., et al. Clinical diagnosis and typing of systemic amyloidosis in subcutaneous fat aspirates by mass spectrometry-based proteomics. Haematologica. 99 (7), 1239-1247 (2014).
- Nakamura, M., et al. Identification of a new transthyretin variant (Ile49) in familial amyloidotic polyneuropathy using electrospray ionization mass spectrometry and nonisotopic RNase cleavage assay. Human Heredity. 49 (4), 186-189 (1999).
- Ueda, M., Ando, Y. Recent advances in transthyretin amyloidosis therapy. Translational Neurodegeneration. 3, 19 (2014).
- Brown, E. E., et al. Genetic testing improves identification of transthyretin amyloid (ATTR) subtype in cardiac amyloidosis. Amyloid. 24 (2), 92-95 (2017).
- Tasaki, M., et al. Rapid detection of wild-type and mutated transthyretins. Annals of Clinical Biochemistry. 53 (4), 508-510 (2015).
- Plante-Bordeneuve, V., et al. Diagnostic pitfalls in sporadic transthyretin familial amyloid polyneuropathy (TTR-FAP). Neurology. 69 (7), 693-698 (2007).
- Hsu, J. L., et al. A prospective, observational study of patients with uncommon distal symmetric painful small-fiber neuropathy. PLoS ONE. 12 (9), 0183948 (2017).
- Salvi, F., Pastorelli, F., Plasmati, R., Bartolomei, I., Dall'Osso, D., Rapezzi, C. Genotypic and phenotypic correlation in an Italian population of hereditary amyloidosis TTR-related (HA-TTR): Clinical and neurophysiological aids to diagnosis and some reflections on misdiagnosis. Amyloid. 19, 58-60 (2012).
- Suhr, O. B., Lundgren, E., Westermark, P. One mutation, two distinct disease variants: Unravelling the impact of transthyretin amyloid fibril composition. Journal of Internal Medicine. 281 (4), 337-347 (2017).
- Vilà-Rico, M., et al. Quantitative analysis of post-translational modifications in human serum transthyretin associated with familial amyloidotic polyneuropathy by targeted LC-MS and intact protein MS. Journal of Proteomics. 127, 234-246 (2015).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone