Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Optical Sectioning and Visualization of the Intervertebral Disc from Embryonic Development to Degeneration
W tym Artykule
Podsumowanie
We present a method to investigate spatial chondrocyte organization in the anulus fibrosus of the intervertebral disc using an optical sectioning method.
Streszczenie
Intervertebral disc (IVD) degeneration is a leading cause of low back pain and it entails a high degree of impairment for the affected individuals. To decode disc degeneration and to be able to develop regenerative approaches a thorough understanding of the cellular biology of the IVD is essential. One aspect of this biology that still remains unanswered is the question of how cells are spatially arranged in a physiological state and during degeneration. The biological properties of the IVD and its availability make this tissue difficult to analyze. The present study investigates spatial chondrocyte organization in the anulus fibrosus from early embryonic development to end-stage degeneration. An optical sectioning method (Apotome) is applied to perform high resolution staining analyses using bovine embryonic tissue as an animal model and human disc tissue obtained from patients undergoing spine surgery. From a very high chondrocyte density in the early embryonic bovine disc, the number of cells decreases during gestation, growth, and maturation. In human discs, an increase in cellular density accompanied the progression of tissue degeneration. As had already been demonstrated in articular cartilage, cluster formation represents a characteristic feature of advanced disc degeneration.
Wprowadzenie
The intervertebral disc (IVD) is a cartilage-based structure that biochemically and with respect to cellular architecture, at first sight, resembles in many ways the articular cartilage1. Indeed, both IVD degeneration and osteoarthritis (OA) of articular cartilage are characterized by joint space narrowing due to cartilage wear, subchondral cyst and osteophyte formation, and subchondral sclerosis2,3. Despite these seeming similarities architecture and functional role of both tissues differ. While the matrix of articular cartilage is mainly formed of an arcade-forming collagen type II network, the IVD consists of three different types of tissue: the collagen type II-rich nucleus pulposus in the center takes up axial loads and transmits them to an encompassing ring of densely packed circular collagen type I fibers which is called anulus fibrosus. Their function is to absorb the translated axial pressures received by the proteoglycan- and water-rich nucleus with their tensile longitudinal fiber strength. At the top and bottom of each nucleus and anulus a hyaline cartilaginous endplate forms the junction to the adjacent vertebrae4 (Figure 1).
In articular cartilage, four distinct spatial chondrocyte patterns can be found: pairs, strings, double strings, small respectively big clusters5,6,7 (Figure 2). Changes in this pattern are associated with OA onset and progression8,9. Spatial chondrocyte organization is also indicative for a direct functional property of cartilage, namely its stiffness, underlining the functional relevance of this image-based grading approach10,11. These patterns can additionally be identified with already existing clinically available technology12. Due to the similarities between the IVD and articular cartilage, it can be hypothesized that characteristic chondrocyte patterns are also present in the IVD. Cluster formation is a phenomenon also observed in the degenerated IVD13,14.
When trying to analyze spatial cellular organization in the IVD, it is necessary to overcome several technical difficulties that are not present when investigating articular cartilage:
First, processing of the tissue itself is much more challenging than with the homogeneous hyaline cartilage which is largely composed of collagen type II. The IVD's main fiber component is collagen type I, which makes it much more difficult to generate thin histologic sections. While in the hyaline articular cartilage even thick sections can easily be analyzed due to the "glass-like" nature of the tissue, the collagen type I network of the IVD is optically highly impenetrable. For this reason, a strong background noise is a common problem in the histology of the IVD. A fast and cheap way to penetrate this optically dense tissue is the use of an optical sectioning device e.g., by means of an Apotome. In such an Apotome, a grid is inserted in the illumination pathway of a conventional fluorescence microscope. In front of the grid a plane-parallel glass plate is placed. This tilts back and forth thus projecting the grid in the image in three different positions. For each z-position, three raw images with the projected grid are created and superimposed. By means of special software, the out of focus light can be calculated out. The underlying principle is that, if the grid is visible, that information is in focus, if not it is considered to be out of focus. With this technique, well focused and high-resolution images can be acquired in a reasonable amount of time.
Secondly, the tissue is hard to come by from human donors. When doing total knee replacement, the entire surface of the joint can be obtained for further analysis during surgery. Although osteoarthritis of a diarthrodial joint is also a disease of the whole joint, there are nevertheless strong focal differences in the quality of the cartilage with usually some areas of the joint still being intact, for example due to reduced loading in that area. This situation is different in the IVD, where surgery is usually only performed when the disc is globally destroyed. When obtaining tissue from human donors from the operation room, the tissue is also highly fragmented and it is necessary to correctly allocate the tissue to one of the three cartilage types of the IVD before doing further analyses. To allow more detailed analyses of also larger tissue sections and to look into the embryonic development of the IVD the choice of an animal model organism is, therefore, necessary.
When choosing such a model organism it is important to have a system which is comparable with the human disc with respect to its anatomy and dimensions, its mechanical loading, the present cell population as well as its tissue composition. For the purpose of the presented technique here we suggest the use of bovine lumbar disc tissue: A critical property of the human disc resulting in its low regenerative potential is the loss of notochordal cells during maturation in the nucleus. However, in numerous model organisms notochordal cells can be detected their entire life long. Most of the few animals which lose their notochordal cells such as sheep, goats or chondrodystrophig dogs have an IVD that is much smaller than human discs. Only lumbar bovine discs present with a comparable sagittal disc diameter to those of human IVDs15.
A key factor leading to early disc degeneration is excessive mechanical loading. The intradiscal pressures of a standing cow in the lumbar spine are around 0.8 MPa with the spine aligned horizontally. Surprisingly these pressures are comparable to the lumbar intradiscal pressures reported for the erect human spine (0.5 MPa)15,16. Also the amount of water and proteoglycans in bovine discs is comparable to that of the IVD from young humans17. Therefore, although the actual movement pattern of the motion segments might differ in quadrupedal animals from the bipedal human, with respect to total loading and disc characteristics, the cow is much closer to human biology than other established animal models for the IVD such as sheep and dogs.
In this protocol we present a technique how to analyze changes in the IVD from the point of view of spatial chondrocyte organization from early embryonic development to end stage degeneration.
Protokół
For the analysis of embryonic development and maturation, bovine discs were used. To evaluate degeneration of the IVD, human samples were analyzed.
Human IVD tissue was obtained from patients undergoing surgery for lumbar disc degeneration, disc prolapse, or spinal trauma in the Department of Orthopaedic Surgery, University Hospital of Tübingen and the BG Trauma Centre Tübingen. Full ethical committee approval was obtained before the commencement of the study (project number 244/2013BO2). Written informed consent was received from all patients before participation. The methods were carried out in accordance with the approved guidelines.
Bovine tissue was obtained from the Bavarian State Office for Health and Food Safety/Oberschleißheim and from a rendering plant in Warthausen (Germany). Local and veterinary authorities' approval was received for tissue from dead animals.
1. Sample harvest
- Human IVD tissue: Place intraoperatively-obtained IVD samples immediately in Dulbecco's Modified Eagle Medium (DMEM) with 2% (v/v) of penicillin-streptomycin and 1.2% of (v/v) amphotericin B. Store at 4 °C until further processing. Process the tissue within 48 h. Alternatively, store the tissues at -20 °C for several weeks.
- Bovine IVD tissue: Ensure to harvest the tissue from the animals within 24 h after death.
Resect the bovine discs with the surrounding vertebrae from the dead animals en-bloc. Transport the frozen tissue on dry ice and store at -20 °C until further processing.
NOTE: If only fluorescence analyses are intended and no further biochemical quantification methods such as ELISA or PCR are planned, perform the tissue fixation as explained below. This allows to keep the tissue longer in storage before it needs to be processed. To prevent deterioration of the tissue matrix, perform fixation within 48 h after harvest unless the tissue is frozen directly.
2. Sample preparation
- Thaw the frozen tissue at room-temperature. Process the tissue as soon as no ice-crystals can be felt any more upon digital gentle compression of the tissue.
NOTE: Perform preparation of the tissue in DMEM in a Petri dish. - Identify the origin of the human IVD tissue (anulus fibrosus, intermediate zone, nucleus pulposus, or cartilaginous endplate) based on macroscopic properties such as collagen density and orientation.
- Take the motion segment consisting of the bovine IVD disc with its two adjacent vertebrae and dissect the disc as a whole from the subchondral bone using a surgical blade (blade number 15).
- Use two anatomic forceps to flip the disc to reach areas more centered. Perform the dissection. Ensure to resect the nucleus pulposus last as it is much thinner than the anulus, prone to tear, and it does not come off in a defined fashion easily.
- Identify the different areas of the cartilage.
- Cut out the area of interest from the whole disc using a surgical blade (blade number 20 or 22). Alternatively, use a cryotome blade.
NOTE: As bovine discs come en-bloc as a part of the spine, the discs can be prepared in-toto. This makes correct identification of the different types of cartilage much easier. When dissecting the disc in the fashion described above, the cartilaginous endplate remains on the vertebrae. If this area shall be investigated, it is best taken off the underlying bone with a chisel working in a slightly bent tangential direction.
- In case that embryos are of a crown-rump length of smaller than 20 cm, ensure to process the embryos in toto to preserve the tissue architecture of the IVD. Do not perform any dissection of the vertebrae in these cases.
- Once the disc has been resected in-toto, identify the different areas of the cartilage.
- Perform the decalcification in ethylenediaminetetraacetic acid (EDTA) (20% (w/v); pH = 7.4) at room temperature. Choose the volume depending on the sample size - the entire tissue must be well covered with EDTA.
- Change the decalcification solution daily which can go for up to 5 days depending on the size of the tissue.
- Verify that the decalcification is successful with a 20 Gauge needle that penetrates the vertebrae without notable resistance.
NOTE: The daily change of decalcification solution is important to prevent the chelate binder EDTA from saturation to maintain reaction effectivity. It also prevents bacterial colonization.
3. Grading of sample age, integrity, and degeneration
- Classify the human disc tissue into one of the five following categories with the help of clinical information as well as X-ray and magnetic resonance imaging18 (Figure 3).
NOTE: Category I: To serve as near-healthy sample use anulus without any radiological sign of IVD degeneration derived from acute spinal trauma.
Category II: To illustrate a situation of acute inflammation with beginning degeneration use tissue from the intermediate zone from patients with clinical symptoms with a duration of less than 4 weeks.
Category III: To describe a situation where the inflammatory reaction had already had time to affect the tissue and cells take tissue from patients who were operated for a nuclear prolapse but with a symptom duration exceeding 4 weeks.
Category IV: For moderate disc degeneration select anulus obtained from surgery with interbody fusion for degenerative disc disease with a Pfirrmann score of 3 or 4 in magnetic resonance imaging19.
Category V: For end stage degeneration process anulus obtained from surgery with interbody fusion for degenerative disc disease with a Pfirrmann score of 5. - Classify the bovine tissue based on the developmental stage/age of the animal into one of the eight categories, as displayed in Table 1.
- Calculate the gestation age on crown-rump length of the embryos based on the formula suggested by Keller:
Gestation age in months =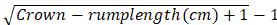
NOTE: Animals in the first 4 weeks of gestation present with a crown-rump length of 0.8-2.2 cm20.
- Calculate the gestation age on crown-rump length of the embryos based on the formula suggested by Keller:
4. Tissue fixation
- Fixate the samples in 10x the volume of the sample of 4% (w/v) formaldehyde solution in phosphate buffered saline (PBS) over night at 4 °C.
NOTE: The formaldehyde solution penetrates tissue at a rate of about 1 mm/hour from each direction. For very small or very big samples an adjustment of the exposure time could be necessary. - Store the tissue in PBS at 4 °C until further processing.
5. Histologic sectioning
- Embed the samples in water-soluble embedding medium on the cryotome knob.
- Place the tissue onto the knob such that either an axial plane is generated or a plane that cuts the collagen type I lamellae perpendicular (e.g., a median sagittal sectioning plane).
NOTE: The tissue has to be fully covered by the embedding medium.
- Place the tissue onto the knob such that either an axial plane is generated or a plane that cuts the collagen type I lamellae perpendicular (e.g., a median sagittal sectioning plane).
- Section the embedded tissue at a thickness of 70 µm in human samples and 40 µm in bovine samples using a standard cryotome.
NOTE: The difference in section thickness is due to the difference in tissue integrity between the intact bovine disc and the highly degenerate human tissue. - Collect the sections on a glass slide.
- Encircle the tissue sections with a hydrophobic pen.
- Rinse the sections 3 times with phosphate-buffered saline (PBS) to remove the water-soluble embedding medium.
6. Fluorescence staining
- Add 60 µL of 1% (v/v) of DAPI (Exmax 358 nm, Emmax 461 nm) as well as 1% (v/v) of Actin Tracking staining (Exmax 540 nm, Emmax 565 nm) in PBS and incubate for 5 min at room temperature.
NOTE: The staining protocol described here is to visualize the nucleus using DAPI nuclear staining (blue) and the cytoplasm using Actin Tracking Stain (red). The IVD has a strong autofluorescence due to the collagen fibers in the green channel. The amount of fluid added to the sections in this protocol is intended for sections of a size of about 5 mm x 5 mm. For larger sections, this amount needs to be increased accordingly. Perform all the works in rooms without direct sunlight exposure and with dimmed lights to prevent bleaching the dye. - Remove the staining fluid with a pipette and wash three times with 60 µL of PBS each time.
- Add a suitable mounting medium and cover the sections with a cover slip.
NOTE: Ensure that there are no air bubbles entrapped when adding the cover slip. This is best done by starting contact of the slip with the slide on one rim and then let the slip come down slowly.
7. Microscopic imaging and processing
- Place a slide with a stained section on the sample holder of the microscope.
NOTE: Due to the dense collagen type I network of the IVD, the scattered light makes the tissue difficult to visualize using conventional fluorescence microscopy. One way to address this problem is to perform optical sectioning using structured illumination. This also allows to render a three-dimensional projection of the entire specimen in both channels (blue and red). This is best done using the structured illumination setting and the Mosaic-mode with a 10x magnification objective lens to obtain an overview of the sample as well as 3D reconstructions of individual patterns. - Start the structured illumination device.
- Perform single field-of-view imaging with a suitable fluorescence microscope, fluorescence filters and adequate illumination.
NOTE: Adjust the exposure time for all the filters used in order to standardize the imaging acquisition. To get an accurate representation of the specimen at a higher resolution image the sections with a higher magnification (e.g., 20x objective). - Postprocess the pictures by optimizing the intensity and brightness using an image optimization software compatible with the fluorescence microscope.
- Perform single field-of-view imaging with a suitable fluorescence microscope, fluorescence filters and adequate illumination.
- To visualize the section as a whole use the mosaic imaging technique
- Open the acquisition settings (press on 6D- Acquisition) from the toolbar panel.
- Adjust the mosaic settings (in MosaiX Register) and define the number of columns and rows of field-of-view images that shall later be merged to one overview image.
- Press Setup and adjust the focus correction of individual tiles.
NOTE: It is almost impossible for a large tissue section to have the whole tissue surface in a single focus plane. Image tiles at different focal levels can be taken by 'MosaiX Acquisition'. - To start the acquisition of the image tiles, press Start.
- Stitch the imaged tiles by using the stitching function (Stitching button) with 20% overlap incorporated in the software.
- Postprocess the pictures by optimizing the intensity and brightness using an image optimization software compatible with the fluorescence microscope.
- To analyze the spatial chondrocyte organization, use the 3D function incorporated in the software.
- Adjust the z-stack settings. Define the scanning parameters: define the start and stop positions in the z-axis and the slice distance by activating Start/Stop button.
NOTE: The software automatically calculates the number of slices. - To start acquisition of the image z-stacks, press Start.
- Postprocess the pictures by optimizing the intensity and brightness using an image optimization software compatible with the fluorescence microscope.
- Adjust the z-stack settings. Define the scanning parameters: define the start and stop positions in the z-axis and the slice distance by activating Start/Stop button.
- Export the pictures with a file format compatible with the image processing software.
NOTE: Export the 3D reconstructions as individual images or/and as an interactive 3D model or in a video format.
8. Cellular pattern identification and density assessment
- Open the exported mosaic pictures of the entire tissue section in an appropriate image processing program.
- Define the areas subjected to cell density assessment by defining regions of interest of 500 µm x 500 µm in the pictures.
NOTE: All tiles that do not have an adequate image quality are excluded from analysis. - Identify individual cellular patterns.
NOTE: Single cells are defined as individual cells - fully encapsulated within the adjacent matrix. Pairs are defined as two adjacent cells in close proximity (<25 µm) whereby the cells are interconnected through their matrixes (see Figure 2). String-formations are at least three chondrocytes aligned in line (from the middle of the nuclei <25 µm). These cells are encompassed by an intact matrix and matrix interconnections can be seen between each cell. Clusters represent multiple cells that are located in direct proximity to each other (<25 µm) and are encapsulated in a large lacuna devoid of matrix. - Use a cell count plug-in for the quantitative analysis of the cellular patterns.
NOTE: The cytoplasm staining represents a verification method to identify the different spatial patterns. - Calculate the cell density by dividing the counted cells by the size of the chosen region of interest.
Wyniki
Using mosaic images, the architecture of the IVD with its dense collagen fiber network in the anulus and the softer nucleus can clearly be recognized (Figure 4). A continuous decrease in cellular density can be observed during embryonic development (Figure 5). While in the early stages of IVD development a cell density of 11,435 cells/mm² in the bovine anulus fibrosus and 17,426 cells/mm² in the bovine nucleus pulposus can be found, these numbers decre...
Dyskusje
Using fluorescence microscopy augmented by mosaic imaging and optical sectioning, we evaluated the spatial arrangement of chondrocytes in the anulus of the lumbar IVD throughout development, maturation, and degeneration. While degenerative tissue could be harvested from patients receiving spine surgery for disc degeneration, analysis of the embryonic period and maturation phase required the use of a model organism (bovine). High cellular densities were noted in the anulus during early embryonic development. In the furthe...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
We thank our co-authors from the original publications for their help and support. We thank Charlotte Emma Bamberger for helping to acquire the apotome images.
Materiały
| Name | Company | Catalog Number | Comments |
| Amphotericin B | Merck KGaA, Germany | A2942 | |
| Adhesion Microscope Slides SuperFrost Plus | R. Langenbrinck, Germany | 03-0060 | |
| ApoTome | Carl Zeiss MicroImaging GmbH, Germany | 462000115 | |
| AxioVision Rel. 4.8 with Modul MosaiX | Carl Zeiss MicroImaging GmbH, Germany | ||
| CellMask Actin Tracking Stain | Thermo Fischer Scientific, US | A57249 | |
| Cryostat | Leica Biosystems, US | CM3050S | |
| DAPI | Thermo Fischer Scientific, US | D1306 | |
| Dulbecco's modified Eagle's medium (DMEM) | Gibco, Life Technologies, Germany | 41966052 | |
| Ethylenediaminetetraacetic acid | Sigma-Aldrich, US | 60004 | |
| Fluorescence Miscoscope - Axio Observer Z1 with Axio Cam MR3 and Colibri | Carl Zeiss MicroImaging GmbH, Germany | 3834000604 | |
| Formaldehyde | Merck KGaA, Germany | 104002 | |
| Image J 1.53a, with Cell counter plugin | National Insittute of Health (NIH), US | ||
| Invitrogen Alexa Fluor 568 Phalloidin | Thermo Fischer Scientific, US | A12380 | |
| Microscopic Cover Glasses | R. Langenbrinck, Germany | 01-1818/1 | |
| PAP Pen Liquid Blocker | Science Sevices GmbH, Germany | N71310 | |
| Penicillin-Streptomycin | Sigma-Aldrich, US | P4333 | |
| Phosphate buffered saline | Sigma-Aldrich,US | P5119 | |
| Scalpel | pf medical AG, Germany | 2023-01 | |
| Tissue-tek O.C.T. Compound | Sakura Finetek, Netherlands | SA6255012 |
Odniesienia
- Urban, J. P. G., Roberts, S. Degeneration of the intervertebral disc. Arthritis Research and Therapy. 5 (3), 120-130 (2003).
- Gupta, K. B., Duryea, J., Weissman, B. N. Radiographic evaluation of osteoarthritis. Radiologic Clinics of North America. 42 (1), 11-41 (2004).
- Pye, S. R., et al. Lumbar disc degeneration: association between osteophytes, end-plate sclerosis and disc space narrowing. Annals of the Rheumatic Diseases. 66 (3), 330-333 (2007).
- Humzah, M. D., Soames, R. W. Human intervertebral disc: structure and function. The Anatomical Record. 220 (4), 337-356 (1988).
- Schumacher, B. L., Su, J. L., Lindley, K. M., Kuettner, K. E., Cole, A. A. Horizontally oriented clusters of multiple chondrons in the superficial zone of ankle, but not knee articular cartilage. The Anatomical Record. 266 (4), 241-248 (2002).
- Rolauffs, B., Williams, J. M., Grodzinsky, A. J., Kuettner, K. E., Cole, A. A. Distinct horizontal patterns in the spatial organization of superficial zone chondrocytes of human joints. Journal of Structural Biology. 162 (2), 335-344 (2008).
- Felka, T., et al. Loss of spatial organization and destruction of the pericellular matrix in early osteoarthritis in vivo and in a novel in vitro methodology. Osteoarthritis and Cartilage. 24 (7), 1200-1209 (2016).
- Rolauffs, B., et al. Onset of preclinical osteoarthritis: the angular spatial organization permits early diagnosis. Arthritis and Rheumatism. 63 (6), 1637-1647 (2011).
- Aicher, W. K., Rolauffs, B. The spatial organization of joint surface chondrocytes: review of its potential roles in tissue functioning, disease and early, preclinical diagnosis of osteoarthritis. Annals of the Rheumatic Diseases. 73 (4), 645-653 (2014).
- Danalache, M., Jacobi, L. F., Schwitalle, M., Hofmann, U. K. Assessment of biomechanical properties of the extracellular and pericellular matrix and their interconnection throughout the course of osteoarthritis. Journal of Biomechanics. 97, 109409 (2019).
- Danalache, M., et al. Changes in stiffness and biochemical composition of the pericellular matrix as a function of spatial chondrocyte organization in osteoarthritic cartilage. Osteoarthritis and Cartilage. 27 (5), 823-832 (2019).
- Tschaikowsky, M., et al. Proof-of-concept for the detection of early osteoarthritis pathology by clinically applicable endomicroscopy and quantitative AI-supported optical biopsy. Osteoarthritis and Cartilage. 29 (2), 269-279 (2021).
- Ciapetti, G., et al. Ex vivo observation of human intervertebral disc tissue and cells isolated from degenerated intervertebral discs. European Spine Journal: Official Publication of the European Spine Society, the European Spinal Deformity Society and the European Section of the Cervical Spine Research Society. 21, 10 (2012).
- Johnson, W. E., Eisenstein, S. M., Roberts, S. Cell cluster formation in degenerate lumbar intervertebral discs is associated with increased disc cell proliferation. Connective Tissue Research. 42 (3), 197-207 (2001).
- Buttermann, G. R., Beaubien, B. P., Saeger, L. C. Mature runt cow lumbar intradiscal pressures and motion segment biomechanics. The Spine Journal: Official Journal of the North American Spine Society. 9 (2), 105-114 (2009).
- Wilke, H. J., Neef, P., Caimi, M., Hoogland, T., Claes, L. E. New in vivo measurements of pressures in the intervertebral disc in daily life. Spine. 24 (8), 755-762 (1999).
- Demers, C. N., Antoniou, J., Mwale, F. Value and limitations of using the bovine tail as a model for the human lumbar spine. Spine. 29 (24), 2793-2799 (2004).
- Hofmann, U. K., et al. Chondrocyte death after mechanically overloading degenerated human intervertebral disk explants is associated with a structurally impaired pericellular matrix. Journal of Tissue Engineering and Regenerative Medicine. 12 (9), 2000-2010 (2018).
- Pfirrmann, C. W., Metzdorf, A., Zanetti, M., Hodler, J., Boos, N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 26 (17), 1873-1878 (2001).
- Habermehl, K. H. . Die Altersbestimmung bei Haus- und Labortieren. , (1975).
- Danalache, M., Erler, A. L., Wolfgart, J. M., Schwitalle, M., Hofmann, U. K. Biochemical changes of the pericellular matrix and spatial chondrocyte organization-Two highly interconnected hallmarks of osteoarthritis. Journal of Orthopaedic Research: Official Publication of the Orthopaedic Research Society. 38 (10), 2170-2180 (2020).
- Bonnaire, F. C., et al. The intervertebral disc from embryonic development to disc degeneration: insights into spatial cellular organization. The Spine Journal: Official Journal of the North American Spine Society. (21), 00198 (2021).
- Vieira-Neto, A., Galvao, K. N., Thatcher, W. W., Santos, J. E. P. Association among gestation length and health, production, and reproduction in Holstein cows and implications for their offspring. Journal of Dairy Science. 100 (4), 3166-3181 (2017).
- Ott, A. Die Entwicklung des schwarzbunten Niederungsrindes von der Geburt bis zum 5. Lebensjahr mit variationsstatistischen Untersuchungen einer Population solcher Rinder von der Geburt bis zum 3. Lebensjahr. Zeitschrift für Tierzüchtung und Züchtungsbiologie. 45 (3), 259-308 (1940).
- Urban, J. P. G., Roberts, S., Ralphs, J. R. The Nucleus of the Intervertebral Disc from Development to Degeneration1. American Zoologist. 40 (1), 53-61 (2000).
- Risbud, M. V., Shapiro, I. M. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nature Reviews. Rheumatology. 10 (1), 44-56 (2014).
- Iatridis, J. C., Michalek, A. J., Purmessur, D., Korecki, C. L. Localized intervertebral disc injury leads to organ level changes in structure, cellularity, and biosynthesis. Cell and Molecular Bioengineering. 2 (3), 437-447 (2009).
- Torre, O. M., Mroz, V., Bartelstein, M. K., Huang, A. H., Iatridis, J. C. Annulus fibrosus cell phenotypes in homeostasis and injury: implications for regenerative strategies. Annals of the New York Academy of Sciences. 1442 (1), 61-78 (2019).
- Rolauffs, B., et al. Proliferative remodeling of the spatial organization of human superficial chondrocytes distant from focal early osteoarthritis. Arthritis and Rheumatism. 62 (2), 489-498 (2010).
- Johnson, W. E., Roberts, S. Rumours of my death may have been greatly exaggerated': a brief review of cell death in human intervertebral disc disease and implications for cell transplantation therapy. Biochemical Society Transactions. 35, 680-682 (2007).
- Roberts, S. Disc morphology in health and disease. Biochemical Society Transactions. 30, 864-869 (2002).
- Lama, P., Kulkarni, J., Tamang, B. The role of cell clusters in intervertebral disc degeneration and its relevance behind repair. Spine Research. 03, 15 (2017).
- Sharp, C. A., Roberts, S., Evans, H., Brown, S. J. Disc cell clusters in pathological human intervertebral discs are associated with increased stress protein immunostaining. European Spine Journal: Official Publication of the European Spine Society, the European Spinal Deformity Society and the European Section of the Cervical Spine Research Society. 18 (11), 1587-1594 (2009).
- Freemont, A. J. The cellular pathobiology of the degenerate intervertebral disc and discogenic back pain. Rheumatology. 48 (1), 5-10 (2009).
- Müllers, Y., et al. Quantitative analysis of F-actin alterations in adherent human mesenchymal stem cells: Influence of slow-freezing and vitrification-based cryopreservation. PLoS One. 14 (1), 0211382 (2019).
- McCann, M. R., Séguin, C. A. Notochord cells in intervertebral disc development and degeneration. Journal of Developmental Biology. 4 (1), 3 (2016).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone