Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Synthesis of Masarimycin, a Small Molecule Inhibitor of Gram-Positive Bacterial Growth
W tym Artykule
Podsumowanie
A detailed protocol is presented for preparing the bacteriostatic diamide masarimycin, a small molecule probe that inhibits the growth of Bacillus subtilis and Streptococcus pneumoniae by targeting cell wall degradation. Its application as a chemical probe is demonstrated in synergy/antagonism assays and morphological studies with B. subtilis and S. pneumoniae.
Streszczenie
Peptidoglycan (PG) in the cell wall of bacteria is a unique macromolecular structure that confers shape, and protection from the surrounding environment. Central to understanding cell growth and division is the knowledge of how PG degradation influences biosynthesis and cell wall assembly. Recently, the metabolic labeling of PG through the introduction of modified sugars or amino acids has been reported. While chemical interrogation of biosynthetic steps with small molecule inhibitors is possible, chemical biology tools to study PG degradation by autolysins are underdeveloped. Bacterial autolysins are a broad class of enzymes that are involved in the tightly coordinated degradation of PG. Here, a detailed protocol is presented for preparing a small molecule probe, masarimycin, which is an inhibitor of N-acetylglucosaminidase LytG in Bacillus subtilis, and cell wall metabolism in Streptococcus pneumoniae. Preparation of the inhibitor via microwave-assisted and classical organic synthesis is provided. Its applicability as a tool to study Gram-positive physiology in biological assays is presented.
Wprowadzenie
Peptidoglycan (PG) is a mesh-like polymer that delineates cell shape and structure in both Gram-positive and Gram-negative bacteria1,2. This heteropolymer is a matrix of amino sugars cross-linked by short peptides3,4,5,6 with a backbone composed of β-(1,4)-linked alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues (Figure 1)1. Attached to the C-3 lactyl moiety of MurNAc is the stem peptide. The metabolism of PG involves a tightly coordinated system of biosynthetic and degradative enzymes to incorporate new material into the cell wall7,8. Degradation of PG is carried out by enzymes collectively referred to as autolysins9 and further classified based on the specificity of the bond cleaved. Autolysins participate in many cellular processes including cell growth, cell division, motility, PG maturation, chemotaxis, protein secretion, genetic competence, differentiation, and pathogenicity10,11. Unraveling the specific biological functions of individual autolysins can be daunting, due in part to functional redundancy. However, recent biophysical8,12,13 and computational studies12 have provided new insight into their roles in PG metabolism. In addition, recent reports have provided further insight into the synthesis14 and membrane-mediated15,16,17 steps in PG metabolism. A thorough understanding of the relationship between degradative and synthetic pathways of PG metabolism could give rise to previously untapped antibiotic targets.
While there have been significant advances in methodology to study glycobiology in eukaryotes, bacterial glycobiology and, in particular, PG metabolism has not advanced at a similar rate. Current chemical approaches to study PG metabolism include fluorescently labeled antibiotics18, fluorescent probes19,20, and metabolic labeling21,22,23,24. These new approaches are providing new ways to interrogate bacterial cell wall metabolism. While some of these strategies are capable of labeling PG in vivo, they can be species-specific19, or only work in strains lacking a particular autolysin25. Many PG labeling strategies are intended for use with isolated cell walls26 or with in vitro reconstituted PG biosynthesis pathways20,27,28. The use of fluorescently labeled antibiotics is currently limited to biosynthetic steps and transpeptidation18.
The current knowledge of bacterial autolysins and their role in cell wall metabolism comes from genetic and in vitro biochemical analysis11,29,30,31,32. While these approaches have provided a wealth of information on this important class of enzymes, deciphering their biological role can be challenging. For instance, due to functional redundancy33, deletion of an autolysin in most cases does not result in halting bacterial growth. This is despite their implied role in cell growth and division7,12. Another complication is that genetic deletion of bacterial autolysins can give rise to meta-phenotypes34. Meta-phenotypes arise from the complex interplay between the pathway affected by the genetic deletion and other interconnected pathways. For instance, a meta-phenotype can arise via a direct effect such as the lack of an enzyme, or an indirect effect such as a disruption of regulators.
Currently, there are only a few inhibitors of glycosidase autolysins such as N-acetylglucosaminidases (GlcNAcase) and N-acetylmuramidases, which can be used as chemical probes to study the degradation of PG. To address this, the diamide masarimycin (previously termed as fgkc) has been identified and characterized35 as a bacteriostatic inhibitor of Bacillus subtilis growth that targets the GlcNAcase LytG32 (Figure 1). LytG is an exo-acting GlcNAcase36, a member of cluster 2 within glycosyl hydrolase family 73 (GH73). It is the major active GlcNAcase during vegetative growth32. To our knowledge, masarimycin is the first inhibitor of a PG-acting GlcNAcase that inhibits cellular growth. Additional studies of masarimycin with Streptococcus pneumoniae found that masarimycin likely inhibits cell wall metabolism in this organism37. Here, the preparation of masarimycin is reported for use as a chemical biology probe to study physiology in the Gram-positive organisms B. subtilis, and S. pneumoniae. Examples of morphological analysis of sub-minimum inhibitory concentration treatment with masarimycin, as well as a synergy/antagonism assay are presented. Synergy and antagonism assays using antibiotics with well-defined modes of action can be a useful way to explore connections between cellular processes38,39,40.
Protokół
1. General methods
NOTE: All compounds were purchased from standard suppliers and used without further purification.
- Carry out thin-layer chromatography (TLC) on an aluminum plate precoated with silica gel XG F254. Detect spots under a UV lamp, by immersion in p-anisaldehyde stain, or by exposing to I2 vapor.
- Record all nuclear magnetic resonance (NMR) spectra on a 400 MHz spectrometer.
NOTE: 1H- NMR and 13C-NMR spectra were referenced to residual solvent peaks. Coupling constants are given in [Hz] and chemical shifts in [ppm]. - Record atmospheric pressure chemical ionization (APCI) mass spectrometry spectra of masarimycin on a compact mass spectrometer equipped with an atmospheric solids analysis probe.
2. General procedure for preparation of masarimycin
NOTE: Perform the below steps in a fume hood.
- Prepare a 0.1 M solution in methanol of each reactant: cyclohexylamine, cyclohexyl carboxaldehyde, o-iodobenzoic acid, and cyclohexyl isocyanide35.
CAUTION: Cyclohexylamine, cyclohexyl isocyanide, and cyclohexyl carboxaldehyde are flammable. They can cause skin corrosion and induce oral, dermal, respiratory, or reproductive toxicity. Keep compounds away from open flames, hot surfaces, and ignition sources. Wear appropriate skin and eye protection, work in a well-ventilated area and avoid inhalation of vapors or mist. For storage, keep bottles tightly closed and store them in a cool, dry place. Store cyclohexyl carboxaldehyde in a desiccator under an N2 atmosphere. - Mix 5 mL of cyclohexylamine (0.1 M solution in methanol) and 5 mL of cyclohexyl carboxaldehyde (0.1 M in methanol) in a capped round bottom flask and stir the solution using a magnetic stir bar on a stir/hot plate for 30 min at 40 °C in a sand bath. Monitor temperature using a thermometer placed approximately 1 cm below the sand surface.
- After 30 min, add 5 mL of cyclohexyl isocyanide (0.1 M solution in methanol) to the solution from step 2.2 and stir for an additional 20 min at 50 °C. Lastly, add 5 mL of o-iodobenzoic acid (0.1 M solution in methanol) to the reaction mixture and continue stirring at 55 °C for 3-5 h.
- Monitor the progress of the reaction periodically by TLC approximately every hour after the above reaction mixture had been stirred for 3 h.
- Cut a 3 cm x 6 cm strip of aluminum-backed TLC plate. Using a #2 pencil, draw a line approximately 1 cm from the bottom. Using a glass microcapillary, spot approximately 5 µL of the reaction mixture onto the TLC plate and allow it to dry.
- To a 150 mL beaker, add enough mobile phase (90:10 hexane: isopropanol) to cover the bottom of the beaker. Using a pair of tweezers, carefully place the above TLC plate into the beaker ensuring that the TLC plate enters the mobile phase evenly. Cover the top of the beaker with a piece of tinfoil.
NOTE: Ensure that the mobile phase does not cover the line and spotted sample. - Allow the mobile phase to travel up the TLC plate until it is approximately 1 cm below the top of the plate. Remove the TLC plate and using a pencil, draw a line indicating the distance traveled by the mobile phase. Allow the TLC plate to dry in a fume hood.
- Once dried, place the TLC plate in a beaker containing a small amount of solid I2 and cover the beaker with a piece of tin foil. Monitor the TLC for the development of yellow/brown spots. Once developed, remove the TLC plate and mark the location of the spots using a pencil (Supplementary Figure 1).
NOTE: If I2 spots are not marked, the stain will dissipate over time. Spots can also be visualized on the TLC plate by UV-light, p-anisaldehyde staining, or potassium permanganate staining (see Supplementary Information). - Calculate Rf values for all visualized spots using the following formula:
Rf =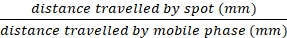
- Consider the reaction complete when only one spot with Rf = 0.3 is visible on the TLC plate. Remove the solvent in a rotatory evaporator under reduced pressure and dry the crude product (obtained as a yellowish-brown oil) under a high vacuum until all methanol is evaporated.
- Dissolve the dried crude product in 30 mL of ethyl acetate and transfer it to a separatory funnel. Extract ethyl acetate sequentially with 1 M HCl (2 x 30 mL), H2O (30 mL), saturated NaHCO3 solution (2 x 30 mL), H2O (30 mL) and saturated NaCl solution (2 x 30 mL). Discard the aqueous layers.
NOTE: The ethyl acetate layer is the top layer in each of the extractions. For each extraction, vigorously shake the separatory funnel containing the ethyl acetate and aqueous solution (HCl, H2O, NaHCO3, or NaCl) and allow the layers to fully separate. - Remove the ethyl acetate layer from the separatory funnel and collect it in an Erlenmeyer flask. Add a spatula full of Na2SO4 (anhydrous) to remove residual water from ethyl acetate.
NOTE: The ethyl acetate solution is considered dry when Na2SO4 in the flask runs freely and does not clump. If Na2SO4 is clumping, an additional spatula of Na2SO4 can be added. - Filter the dried ethyl acetate solution through #1 filter paper to remove Na2SO4. Wash the filter paper with a small amount of ethyl acetate. Place the filtered ethyl acetate solution in a round bottom flask and remove the solvent on a rotatory evaporator under reduced pressure to obtain masarimycin as oil once all the ethyl acetate is removed.
- Dissolve the masarimycin oil obtained above in a minimal amount (1-2 mL) of 9:1 hexane: isopropanol and stir on a magnetic stir plate until all the compound is dissolved.
- Purify the dissolved masarimycin by flash chromatography using a 12 g normal phase silica flash column.
- Equilibrate the flash column with 10 column volumes of mobile phase (99:1 hexane: isopropanol) with the instrument set at a flow rate of 15 mL/min.
NOTE: After equilibration is completed, stop the flow and disconnect the top of the column from the system. - Draw up dissolved masarimycin using a 5 mL syringe. Connect the syringe directly to the top of the equilibrated flash column and inject the solution into the column. Reconnect the loaded column to the flash chromatography system and initiate the gradient elution.
- Elute masarimycin from the column using gradient elution to a final mobile phase concentration of 10:90 hexane: isopropanol over 12 column volumes. Monitor the elution of masarimycin via absorption at 230 and 254 nm.
- Collect the compounds eluted from the column by a fraction collector that collects 20 mL of solvent per fraction.
NOTE: If a flash chromatography system is not available, purification of masarimycin can be performed via a gravity silica column with a 3:1 (hexane: ethyl acetate) mobile phase. Fractions containing masarimycin can be identified by TLC using the same mobile phase. Visualization of TLC spots was done with either UV light, I2 vapor, or potassium permanganate staining. - Identify fractions containing masarimycin by TLC (steps 2.5-2.9) or mass spectrometry on a compact mass spectrometer equipped with an atmospheric solids analysis probe. Dry the final product under vacuum (~0.3 mbar).
NOTE: Masarimycin is routinely obtained as a colorless oil or solid with a yield of 55%-70% with respect to mmol of cyclohexyl carboxaldehyde added to the reaction. Calculate the final yield of masarimycin by obtaining the mass of the purified masarimycin and calculating the theoretical yield of the reaction using the following formula:
% yield = x 100%
x 100%
- Equilibrate the flash column with 10 column volumes of mobile phase (99:1 hexane: isopropanol) with the instrument set at a flow rate of 15 mL/min.
- Confirm the structure of masarimycin by NMR.
- Dissolve ~10 mg of masarimycin sample in 0.5 mL of CDCl3. Using a Pasteur pipet, transfer the solution to a 5 mm NMR tube and cap the tube. Place the NMR tube in the spectrometer.
- Acquire 1H and 13C NMR spectra using manufacturer preset experiments. Chemical shift assignments and representative spectra are provided in Supplementary Figures 3-4.
- Store masarimycin dry or dissolved in DMSO (25 mM final concentration) at -20 °C until use.
3. Microwave procedure for preparation of masarimycin
- Prepare 0.6 M solutions of cyclohexylamine, cyclohexyl carboxaldehyde, cyclohexyl isocyanide, and o-iodobenzoic acid in acetonitrile.
- Add a stir bar and 10 mL of acetonitrile to a glass microwave reaction vial.
- Add 2 mL of cyclohexylamine (0.6 M in acetonitrile), 2 mL of cyclohexyl carboxaldehyde (0.6 M in acetonitrile), and 7 mL of acetonitrile to the vial.
- Place the microwave reaction vial in the microwave carousel. Stir the mixture, heat it for 30 min at 50°C at a power setting of 400 W, and allow it to cool to room temperature.
- Add 2 mL of o-iodobenzoic acid (0.6 M in methanol) and 2 mL of cyclohexyl isocyanide (0.6 M in acetonitrile) to the vial. Stir the mixture, heat it to 100 °C in the microwave for 40 min at a power setting of 400 W and allow it to cool to room temperature.
- Monitor the progress of the reaction by TLC (90:10 hexane: isopropanol) using I2 vapor after the completion of step 3.5.
NOTE: If TLC shows that the reaction is incomplete (i.e., multiple spots on TLC), place the reaction vial back in the microwave and set the microwave conditions described in step 3.5. - Once the reaction is complete, pour the solution into a 100 mL round-bottom flask and evaporate it to dryness using a rotary evaporator.
- Follow steps 2.6-2.16 above to complete the aqueous workup, purification, and characterization of masarimycin.
4. Synergy and antagonism assay
- Grow Streptococcus pneumoniae R6 on Mueller-Hinton (MH) agar plates containing 5% (v/v) sheep blood at 37 °C under anaerobic conditions. In all experiments, use second passage cells grown in 5 mL of MH broth at 37 °C under anaerobic conditions until OD600 is ~0.4.
- Subject the inhibitors masarimycin and optochin to serial 1:2 dilutions in respective solvents, with the resulting concentrations flanking the minimum inhibitory concentration (MIC) values of each inhibitor.
- Make the initial dilution of masarimycin in dimethyl sulfoxide (DMSO) until a concentration of 100 µM is reached. From this point, make masarimycin dilutions in MH broth. Prepare optochin stock solution (3.5 mM) by dissolving commercially available optochin (see Table of Materials) in sterile MH broth.
NOTE: Masarimycin stock solutions were made at 25 mM in DMSO.
- Make the initial dilution of masarimycin in dimethyl sulfoxide (DMSO) until a concentration of 100 µM is reached. From this point, make masarimycin dilutions in MH broth. Prepare optochin stock solution (3.5 mM) by dissolving commercially available optochin (see Table of Materials) in sterile MH broth.
- To a sterile 96-well microtitre plate, add 2 µL aliquots of each optochin dilution to each row of the plate. To the same plate, add 2 µL aliquots of each masarimycin dilution to each column to create an array of optochin and masarimycin concentrations on the plate (Figure 2).
- Add sterile MH broth (93 µL) to each well containing the above inhibitors. Inoculate the microtitre plates with 5 µL of culture (OD600 ~0.4) from step 4.1.
NOTE: Inoculation of the 96-well plate is typically done under anaerobic conditions in an anaerobic workstation. The final volume in the well is 100 µL. - Grow cultures for 18 h at 37 °C under anaerobic conditions, followed by the addition of 30 µL of 0.01% (m/v) solution of resazurin sodium salt. Incubate the plate at room temperature for 15 min to allow the formation and stabilization of color.
NOTE: Resazurin solution is prepared by dissolving the compound in distilled water and can be stored at 4 °C for up to two weeks. - Directly read the concentration values from the plate and assign the lowest inhibitor concentration for which no bacterial growth is observed (blue color) as [X] (see step 4.7.1), i.e., the lowest inhibitory concentration of the drug in the presence of the co-drug.
NOTE: Positive bacterial growth is identified in the wells by the resazurin dye turning pink. MIC values for each drug alone (i.e., in the absence of co-drug) are determined in a similar manner using the resazurin MIC assay35 with each drug separately (Supplemental Figure 5). MICs in S. pneumoniae are 7.8 µM and 15.85 µM for masarimycin and optochin, respectively. - Determine the fractional inhibitory concentration (FIC) and FIC index (FICI) using the following equations.
- FIC= [X]/MICx, where [X] (from step 4.6) is the lowest inhibitory concentration of the drug in the presence of the co-drug, and MICx is the lowest inhibitory concentration of the drug in the absence of the co-drug.
- FICI= FICmasarimycin + FICantibiotic
NOTE: FICI < 0.5 = synergistic, 0.5 < FICI < 1 = additive, 1 < FICI < 4 = indifferent, FICI > 4 = antagonistic.
5. Morphological study
- Grow Bacillus subtilis 11774 on Luria-Bertani (LB) agar plates (10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl) containing 1.5% Bacto agar at 37 °C. In all experiments, use second passage cells grown in 5 mL of LB broth at 37 °C until OD600 = 1. Grow S.pneumoniae in the same manner as in step 4.1.
- After obtaining a cell culture density with OD600nm = 1 for B. subtilis, or OD600nm = 0.4 for S. pneumoniae, add masarimycin using a pipette to the culture tube labeled "treated" to a final concentration of 3.8 µM (0.75x MIC for B.subtilis), or 5.85 µM (0.75x MIC for S.pneumoniae). To the second culture tube labeled "control", add an equivalent volume of DMSO.
- For B.subtilis, place the samples in an incubator at 37 °C for 90 min with shaking at 150 rpm. For S. pneumoniae, incubate the cells without shaking under anaerobic conditions.
- After 90 min, chemically fix the cultures in a 1:10 mixture (v/v) of culture media and fixing buffer (20 mM HEPES, 1% formaldehyde (pH 6.8)) at 4 °C overnight. After fixing is complete, apply 10-20 µL of samples to glass microscope slides using a pipette and allow them to air dry. Fix the air-dried samples by heating the glass slides using a Bunsen burner.
- After heat-fixing, stain samples with the addition of 100 µL of 0.1% (m/v) methylene blue (solution in 20% (v/v) ethanol). Incubate the stained slides for 10 min and wash away the excess dye with dH2O. Then, gently heat the stained slides to 60 °C in an oven for 15-20 min to bring cells to a common focal plane.
- Seal the stained samples by placing a microscope coverslip over the stained cells. Then, seal the edges using microscope slide cement. Place the sealed microscope slide on the microscope stage and bring the image into focus at 100x magnification using bright-field microscopy.
- Place a drop of immersion oil on the microscope slide and bring the field of view to focus using 1000x magnification. Acquire micrographs using a camera attached to the microscope and its associated software. Acquire images using the auto white balance and aperture settings on the software.
NOTE: Alternatively, images can be processed using the open-source ImageJ software.
Wyniki
Masarimycin is a small molecule bacteriostatic inhibitor of B. subtilis and S. pneumoniae and has been shown to inhibit the exo-acting GlcNAcase LytG in B. subtilis35,37 and target the cell wall in S. pneumoniae37. Masarimycin can be efficiently prepared either by the classical or microwave-assisted organic synthesis with yields in the 55%-70% range. Microwave-assisted synthesis has the advantage of a si...
Dyskusje
Masarimycin is a single micromolar bacteriostatic inhibitor of B. subtilis35 and S. pneumoniae37 growth. In B. subtilis, masarimycin has been shown to inhibit the GlcNAcase LytG35, while the precise molecular target in the cell wall of S. pneumoniae has not been identified37. Synthesis of masarimycin using either the classical organic synthesis or microwave procedure provides the inhibitor in good yi...
Ujawnienia
Reid, C. W. has intellectual property involving specific applications of masarimycin.
Podziękowania
Research was supported by the National Science Foundation under grant number 2009522. NMR analysis of masarimycin was supported by the National Science Foundation major research instrumentation program award under grant number 1919644. Any opinions, findings, and conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Materiały
| Name | Company | Catalog Number | Comments |
| 2-Iodobenzoic acid | SIGMA-ALDRICH | I7675-25G | corrosive, irritant, light yellow to orange-brown powder |
| 2-Propanol | SIGMA-ALDRICH | 109827-4L | flammable, irritant, colorless liquid |
| Acetonitrile | SIGMA-ALDRICH | 34851-4L | flammable, irritant, colorless liquid |
| Aluminum backed silica plates | Sorbtech | 4434126 | silica gel XG F254 on aluminum backed plates |
| chloroform-d | SIGMA-ALDRICH | 151823-50G | solvent for NMR |
| Compact Mass Spectrometer | Advion-Interchim | Advion CMS | compact mass spectrometer equiped with APCI source and atmospheric solids analysis probe |
| Corning Costar 96 well flat bottom plates-sterile | fisher chemical | 07-200-90 | for synergy/antagonism assays |
| cover slips | fisher chemical | 12-547 | for microscopy |
| Cyclohexanecarboxaldehyde | CHEM-IMPEX INT'L INC. | 24451 | flammable, irritant, colorless to pink liquid |
| Cyclohexyl isocyanide | SIGMA-ALDRICH | 133302-5G | irritant, colorless liquid, extremly unpleasant odor |
| Cyclohexylamine | SIGMA-ALDRICH | 240648-100ML | corrosive, flammable, irritant, colorless liquid unless contaminated |
| Ethyl acetate | SIGMA-ALDRICH | 537446-4L | flammable, irritant, colorless liquid |
| flash silica cartridge (12g) | Advion-Interchim | PF-50SIHP-F0012 | pack of flash silica columns (12g) for purification of masarimycin |
| formaldehyde | SIGMA-ALDRICH | F8775-25ML | fixing agent for microscopy |
| HEPES | SIGMA-ALDRICH | H8651-25G | buffer for microscopy fixing solution |
| Hexane, mixture of isomers | SIGMA-ALDRICH | 178918-4L | environmentally damaging, flammable, irritant, health hazard, colorless liquid |
| High performance compact mass spectrometer | Advion | expression | Atmospheric Solids Analysis Probe (ASAP), low resolution |
| High Vac | eppendorf | Vacufuge plus | vacuum aided by centrifugal force and temperature |
| Hydrochloric acid | SIGMA-ALDRICH | 258148-2.5L | corrosive, irritant, colorless liquid |
| hydrochloric acid | SIGMA-ALDRICH | 320331-2.5L | strong acid |
| immersion oil | fisher chemical | 12-365-19 | for microscopy |
| Iodine, resublimed crystals | Alfa Aesar | 41955 | environmentally damaging, irritant, health hazard, dark grey/purple crystals |
| Mestre Mnova | MestreLab Research | software for processing NMR spectra | |
| Methanol | SIGMA-ALDRICH | 439193-4L | flammable, toxic, health hazard, colorless liquid |
| methylene blue | SIGMA-ALDRICH | M9140-25G | microscopy stain for staining cell walls |
| meuller-hinton agar plates + 5% sheep blood | fisher chemical | B21176X | growth media for Streptococcus pneumoniae |
| meuller-hinton broth | fisher chemical | DF0757-17-6 | growth media for Streptococcus pneumoniae |
| microscope slides | fisher chemical | 22-310397 | for microscopy |
| Microwave Synthesis Labstation | MILESTONE | START SYNTH | device that requires the ventilation of a fume hood, equipped with synthesis carousel |
| NMR tubes | SIGMA-ALDRICH | Z562769-5EA | 5mm NMR tubes 600 MHz |
| Nuclear Magnetic Resonance (NMR) | Bruker | Ascend 400 | large superconducting magnet (400MHz) |
| optochin | fisher chemical | AAB21627MC | ethylhydrocupreine hydrochloride |
| petrie plates | Celltreat | 229695 | for preparing agar plates for bacterial growth |
| Primo Star Bright field/Phase contrast Microscope with ERc5s camera | Zeiss | for morphology studies | |
| puriFlash | interchim | XS520plus | flash chromatography purification system |
| resazurin | SIGMA-ALDRICH | R7017-1G | for synergy/antagonism assays |
| Rotary Evaporator | Heidolph | Hei-VAP Value "The Collegiate" | solvent evaporator |
| Sodium bicarbonate | SIGMA-ALDRICH | S6014-500G | irritant, white powder |
| Sodium chloride | fisher chemical | S271-1 | crystalline, colorless |
| Sodium chloride | SIGMA-ALDRICH | S5886-500G | for growth of B.subtilis and preparation of LB media |
| Sodium sulfate | SIGMA-ALDRICH | 7985592-500G | anhydrous, granular, white |
| tryptone | fisher chemical | BP1421-500 | for growth of B.subtilis and preparation of LB media |
| Whitney DG250 Workstation | Microbiology International | DG250 | anaerobic workstation. Anaerobic gas mixture used: 5% hydrogen, 10% carbon dioxide, balance nitrogen |
| yeast extract | fisher chemical | BP1422-500 | for growth of B.subtilis and preparation of LB media |
| Zen Lite (blue) software | Zeiss | for acquiring micrographs |
Odniesienia
- Vollmer, W., Blanot, D., de Pedro, M. A. Peptidoglycan structure and architecture. FEMS Microbiology Review. 32 (2), 149-167 (2008).
- Munita, J. M., Bayer, A. S., Arias, C. A. Evolving resistance among Gram-positive pathogens. Clinical Infectious Diseases. 61, 48-57 (2015).
- Vollmer, W., Bertsche, U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochimica Biophysica Acta. 1778 (9), 1714-1734 (2008).
- Vollmer, W., Höltje, J. -. V. The architecture of the murein (peptidoglycan) in Gram-negative bacteria: vertical scaffold or horizontal layer(s). Journal of Bacteriology. 186 (18), 5978-5987 (2004).
- Clarke, A. J. Compositional analysis of peptidoglycan by high-performance anion-exchange chromatography. Analytical Biochemistry. 212 (2), 344-350 (1993).
- Kim, S. J., Chang, J., Singh, M. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochimica Biophysica Acta. 1848, 350-362 (2014).
- Koch, A. L., Doyle, R. J. Inside-to-outside growth and turnover of the wall of gram-positive rods. Journal of Theoretical Biology. 117 (1), 137-157 (1985).
- Beeby, M., Gumbart, J. C., Roux, B., Jensen, G. J. Architecture and assembly of the Gram-positive cell wall. Molecular Microbiology. 88 (4), 664-672 (2013).
- Shockman, G. D., Daneo-Moore, L., Kariyama, R., Massidda, O. Bacterial walls, peptidoglycan hydrolases, autolysins, and autolysis. Microbial Drug Resistance. 2 (1), 95-98 (1996).
- Dijkstra, A. J., Keck, W. Peptidoglycan as a barrier to transenvelope transport. Journal of Bacteriology. 178 (19), 5555-5562 (1996).
- Blackman, S. A., Smith, T. J., Foster, S. J. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology. 144, 73-82 (1998).
- Misra, G., Rojas, E. R., Gopinathan, A., Huang, K. C. Mechanical consequences of cell-wall turnover in the elongation of a Gram-positive bacterium. Biophysical Journal. 104 (11), 2342-2352 (2013).
- Wheeler, R., et al. Bacterial cell enlargement requires control of cell wall stiffness mediated by peptidoglycan hydrolases. mBio. 6 (4), 00660 (2015).
- Taguchi, A., Kahne, D., Walker, S. Chemical tools to characterize peptidoglycan synthases. Current Opinion in Chemical Biology. 53, 44-50 (2019).
- Welsh, M. A., Schaefer, K., Taguchi, A., Kahne, D., Walker, S. Direction of chain growth and substrate preferences of shape, elongation, division, and sporulation-family peptidoglycan glycosyltransferases. Journal of the American Chemial Society. 141 (33), 12994-12997 (2019).
- Rubino, F. A., et al. Detection of transport intermediates in the peptidoglycan flippase MurJ identifies residues essential for conformational cycling. Journal of the American Chemical Society. 142 (12), 5482-5486 (2020).
- Sjodt, M., et al. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature. 556 (7699), 118-121 (2018).
- Tiyanont, K., et al. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proceedings of the National Academy of Science U S A. 103 (29), 11033-11038 (2006).
- Lebar, M. D., et al. Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. Journal of the American Chemical Society. 136 (31), 10874-10877 (2014).
- Do, T., Page, J. E., Walker, S. Uncovering the activities, biological roles, and regulation of bacterial cell wall hydrolases and tailoring enzymes. Journal of Biological Chemistry. 295 (10), 3347-3361 (2020).
- Liang, H., et al. Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nature Communications. 8, 15015 (2017).
- DeMeester, K. E., et al. Metabolic incorporation of N-acetyl muramic acid probes into bacterial peptidoglycan. Current Protocol in Chemical Biology. 11 (4), 74 (2019).
- Lazor, K. M., et al. Use of Bioorthogonal N-acetylcysteamine (SNAc) analogues and peptidoglycan O-acetyltransferase B (PatB) to label peptidoglycan. The FASEB Journal. 32, 630 (2018).
- Wang, Y., Leimkuhler-Grimes, C. Fluorescent labeling of the carbohydrate backbone of peptidoglycan to track degradation in vivo. The FASEB Journal. 29, (2015).
- Kuru, E., et al. In probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angewandte Chemie International Edition. 51 (50), 12519-12523 (2012).
- Zhou, R., Chen, S., Recsei, P. A dye release assay for determination of lysostaphin activity. Analytical Biochemistry. 171 (1), 141-144 (1988).
- Qiao, Y., et al. Lipid II overproduction allows direct assay of transpeptidase inhibition by β-lactams. Nature Chemical Biology. 13 (7), 793-798 (2017).
- Lebar, M. D., et al. Forming cross-linked peptidoglycan from synthetic Gram-negative lipid II. Journal of the American Chemical Society. 135 (12), 4632-4635 (2013).
- Chen, R., Guttenplan, S. B., Blair, K. M., Kearns, D. B. Role of the D-dependent autolysins in Bacillus subtilis population heterogeneity. Journal of Bacteriology. 191 (18), 5775-5784 (2009).
- Yukie, S., Miki, K., Yoshio, N., Kuniaki, T., Yoshihisa, Y. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infection and Immunity. 73 (6), 3512-3520 (2005).
- Domenech, M., García, E., Moscoso, M. In vitro destruction of Streptococcus pneumoniae biofilms with bacterial and phage peptidoglycan hydrolases. Antimicrobial Agents and Chemotherapy. 55 (9), 4144-4148 (2011).
- Horsburgh, G. J., Atrih, A., Williamson, M. P., Foster, S. J. LytG of Bacillus subtilis is a novel peptidoglycan hydrolase: the major active glucosaminidase. Biochemistry. 42 (2), 257-264 (2003).
- Vermassen, A., et al. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Frontiers in Microbiology. 10, 331 (2019).
- Martin-Galiano, A. J., Yuste, J., Cercenado, M. I., de la Campa, A. G. Inspecting the potential physiological and biomedical value of 44 conserved uncharacterised proteins of Streptococcus pneumoniae. BMC Genomics. 15, 652 (2014).
- Nayyab, S., et al. Diamide inhibitors of the Bacillus subtilis N-acetylglucosaminidase LytG that exhibit antibacterial activity. ACS Infectioius Diseases. 3 (6), 421-427 (2017).
- Lipski, A., et al. Structural and biochemical characterization of the β-N-acetylglucosaminidase from Thermotoga maritima: Toward rationalization of mechanistic knowledge in the GH73 family. Glycobiology. 25 (3), 319-330 (2014).
- Haubrich, B. A., et al. Inhibition of Streptococcus pneumoniae autolysins highlight distinct differences between chemical and genetic inactivation. bioRxiv. , 300541 (2020).
- Farha, M. A., et al. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chemical Biology. 8 (1), 226-233 (2013).
- Lehár, J., et al. Chemical combination effects predict connectivity in biological systems. Molecular Systems Biology. 3 (1), 80 (2007).
- Farha, M. A., et al. Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase. Proceedings of the National Academy of Science U S A. 112 (35), 11048-11053 (2015).
- Palomino, J. C., et al. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy. 46 (8), 2720-2722 (2002).
- Odds, F. C. Synergy, antagonism, and what the chequerboard puts between them. Journal of Antimicrobial Chemotherapy. 52 (1), 1 (2003).
- Arrigucci, R., Pozzi, G. Identification of the chain-dispersing peptidoglycan hydrolase LytB of Streptococcus gordonii. PLoS One. 12 (4), 0176117 (2017).
- Bai, X. -. H., et al. Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis. of Biological Chemistry. 289 (34), 23403-23416 (2014).
- Garcia, P., Gonzalez, M. P., Garcia, E., Lopez, R., Garcia, J. L. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Molecular Microbiology. 31 (4), 1275-1281 (1999).
- Giladi, M., Altman-Price, N., Levin, I., Levy, L., Mevarech, M. FolM, a new chromosomally encoded dihydrofolate reductase in Escherichia coli. Journal of Bacteriology. 185 (23), 7015-7018 (2003).
- Chua, P. R., et al. Effective killing of the human pathogen Candida albicans by a specific inhibitor of non-essential mitotic kinesin Kip1p. Molecular Microbiology. 65 (2), 347-362 (2007).
- Rico-Lastres, P., et al. Substrate recognition and catalysis by LytB, a pneumococcal peptidoglycan hydrolase involved in virulence. Scientific Reports. 5, 16198 (2015).
- Vollmer, W., et al. The cell wall of Streptococcus pneumoniae. Microbiology Spectrum. 7 (3), (2019).
- Massidda, O., Nováková, L., Vollmer, W. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division. Environmental Microbiology. 15 (12), 3133-3157 (2013).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone