A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Assessing Two-dimensional Crystallization Trials of Small Membrane Proteins for Structural Biology Studies by Electron Crystallography
* These authors contributed equally
In This Article
Summary
Evaluating two-dimensional (2D) crystallization trials for the formation of ordered membrane protein arrays is a highly critical and difficult task in electron crystallography. Here we describe our approach in screening for and identifying 2D crystals of predominantly small membrane proteins in the range of 15 – 90kDa.
Abstract
Electron crystallography has evolved as a method that can be used either alternatively or in combination with three-dimensional crystallization and X-ray crystallography to study structure-function questions of membrane proteins, as well as soluble proteins. Screening for two-dimensional (2D) crystals by transmission electron microscopy (EM) is the critical step in finding, optimizing, and selecting samples for high-resolution data collection by cryo-EM. Here we describe the fundamental steps in identifying both large and ordered, as well as small 2D arrays, that can potentially supply critical information for optimization of crystallization conditions.
By working with different magnifications at the EM, data on a range of critical parameters is obtained. Lower magnification supplies valuable data on the morphology and membrane size. At higher magnifications, possible order and 2D crystal dimensions are determined. In this context, it is described how CCD cameras and online-Fourier Transforms are used at higher magnifications to assess proteoliposomes for order and size.
While 2D crystals of membrane proteins are most commonly grown by reconstitution by dialysis, the screening technique is equally applicable for crystals produced with the help of monolayers, native 2D crystals, and ordered arrays of soluble proteins. In addition, the methods described here are applicable to the screening for 2D crystals of even smaller as well as larger membrane proteins, where smaller proteins require the same amount of care in identification as our examples and the lattice of larger proteins might be more easily identifiable at earlier stages of the screening.
Protocol
1. Grid Preparation of 2D Crystallization Trials
- Carbon-coated 400-mesh copper EM grids are prepared by negative stain. Uranyl acetate is frequently used and provides a long-lasting stain in terms of storage of the solution for several months before use as well as suitability for long-term storage of grids. In contrast, other negative stains such as uranyl formate, while providing excellent staining, need to be freshly made 1. For fast preparation of a large number of grids to be used for screening of 2D crystallization trials, a modified version of negative staining is used. A volume of 2 μL of sample is pipetted onto a carbon-covered EM grid and incubated for 60 s. This is followed by blotting from the edge with a torn piece of Whatman #4 filter paper (Figure 1; video), and then 2 μL of 1% uranyl acetate are applied immediately, which are again blotted from the edge of the grid after 30 s. Touching the grid on the rim with the torn edge of the filter paper ensures optimal removal of liquid without removal of proteoliposomes. Furthermore, drying at the edge of the grid ensures improved preservation of the carbon film. Care must be taken in preparation and handling of the grids, as breakage of the delicate carbon film prevents sample adhesion and can result in an inaccurate representation of the sample. While traditionally larger sample volumes of 5 μL were and are routinely used for grid preparation, precious samples can be saved by reducing the volume to 2 μL or less 2.
- To produce the largest possible membranes, some of our samples require high concentrations of glycerol or sucrose (10-20%) to be present in the dialysis buffer. This can have a negative effect on grid preparation, as glycerol or sucrose is highly viscous and prevents uranyl acetate from properly penetrating the buffer, and thus incompletely staining the membranes. Consequently a possible lattice will be obscured. Either the buffer can be exchanged by centrifugation of the samples and replacement by glycerol/sucrose-free buffer (not shown), or the grids can be washed with buffer or glycerol/sucrose-free buffer in one to several cycles before negative staining similar to a technique used for cryo-EM 3,4.
2. Assessing 2D Crystallization Trials by EM
- Depending on the type of sample holder, either one or multiple grids are loaded. A magnification of 2-10K, which will be referred to here as intermediate magnification, allows for a first impression of average distribution and extent of dispersion of membranes, morphology, and size, which is taken note of in the laboratory notebook 5. Suitable areas are recorded as representative overviews with the help of a CCD camera or, if a CCD camera is not available, on film.
- Low magnification in the range of roughly 400-800x is used at times when an overview of the entire sample/grid is desired. While not employed with every grid, low magnification gives valuable information of help in the evaluation of the grid preparation in terms of several aspects: both negative staining and potential partial breakage of carbon film, sample concentration on the grid, and possibly uneven proteoliposome distribution. Individual grid squares might be viewed with the binoculars or CCD camera. With some EMs it is possible to save positions of particular interest, which can be recalled for later inspection at higher magnifications.
- Once a grid area of interest has been identified at either low or intermediate magnification, the magnification is changed to approximately 50K-60K. Depending on membrane, crystal and unit cell size, if known, magnifications as low as 30K and as high as 80K are used. The magnification range of 30-80K will be referred to as high magnification for the purpose of screening for 2D crystals. Focusing occurs either in the focus setting of the low-dose set-up, with subsequent switch to image/photo setting, or in the vicinity of the area of interest.
- In cases of ambiguity in whether the area of interest is indeed a membrane, the sample is inspected for carbon-film in the size of proteoliposomes, mica, or other artifacts. For this purpose, edges reveal typical folding and morphology.
- Now the area of interest is inspected with a CCD camera. A CCD image is collected at 30K-80K magnification, depending on membrane size, protein or unit cell size, or known crystalline area. The lattice of a smaller and/or mostly hydrophobic membrane protein is not necessarily visible by visual assessment of the CCD image itself. Either the entire image is used for an online-Fourier transform (FT, or fast FT -FFT). This FT will contain a significant amount of noise, however, if the ordered array is small. Thus, a reduced boxed image size will allow for an improved signal-to-noise ratio of a smaller crystal and easier identification. For this purpose, the box is moved over the image and a live FT is evaluated.
The intensity/brightness of the beam is adjusted keeping low-dose conditions for the sample, as well as CCD camera settings in mind. Depending on the CCD camera used, the gamma of the live FT is adjusted for optimal identification of ordered arrays. An overly high value can obscure spots due to noise contributions, and an excessively low gamma value will prevent weaker spots from being identified. These weaker spots might be due to smaller crystalline arrays with spots in the FT barely above the noise level.
While data at higher resolution has been collected of a small number of samples 6, commonly the resolution of negatively stained 2D crystals is limited to or should not be expected to be better than roughly 15Å resolution. With a defocus of approximately -400 nm, not more than 1-3 orders of spots are expected to be easily identified. Samples are generally not evaluated for resolution, as cryo-EM data collection will give a proper indication of highest achievable resolution. Sharpness of spots and possible mosaicity are noted though.
- Different membranes, as well as membrane morphologies, are evaluated for order at high magnification. This is particularly critical at beginning or intermediate stages of 2D crystallization trials, as smaller rather than bigger proteoliposomes or membrane patches can contain the most promising areas. Very low percentages of 2D crystals will require image acquisition and FT, or optical diffraction, of a large number of images since initial identification of ordered arrays will frequently lead to rapid improvement of size and quality 2,4,7.
3. Representative Results
Ideally ordered proteoliposomes display readily recognizable, sharp spots. Large and well-ordered crystals are easily identified by online-FT of CCD images or optical diffraction of micrographs.
The example shows 2D crystals of a small membrane protein of 18kDa that are up to several microns in size. Spots on the FT are easily identified and sharp. The movement of the live-FT box shows that the lattice is continuous without mosaicity. The lattice of a larger protein with a more extensive soluble domain can be identified on the small screen of the EM. CCD image collection and FT is necessary to provide a means of better assessment and to gain information on, e.g., possible mosaicity (show FT). When calculating an FT of a proteoliposome that is not ordered, noise can initially be mistaken for spots. While the box for the live-FT is moved, however, the spots will disappear. On the other hand, small arrays, with questionable crystallinity, will have their spots remaining stationary when the live-FT is moved even slightly over the image area. Furthermore, these small crystals can be recognized by usually having the same unit cell sizes, and distances between spots in different FTs can be measured in various ways, such as with a circle of a specific size. Lipid crystals display a distinct lattice morphology and FT.
It is not uncommon to encounter precipitation in initial trials. Here protein precipitation without reconstitution needs to be distinguished from small lipid aggregates though. Samples that appear to be precipitates at low magnification frequently turn out to be lipid aggregates when viewed at higher magnification. Upon inspection at 30-50K, the edges of these dark structures reveal them to be composed of membranes with no precipitation of the protein. These are important observations as the lipid aggregates might be increased in size to large membranes in the following experiments.
Poor results in evaluating samples are sometimes connected to a low membrane concentration that prevents proper and fast screening. This can frequently be overcome with the use of a higher protein concentration for 2D crystallization by dialysis. Alternatively, the membranes can be left to settle for a few days at the bottom of the Eppendorf tube during storage. In some cases rapid, or almost instant settling of membranes occurs and pipetting from the bottom of the tube will result in a higher membrane density on the grid. Another much faster option is centrifugation (at 3000-8000 rpm for 1-3 minutes) of samples with subsequent sampling from the bottom of the tube.
Samples under optimal conditions will contain a large percentage of 2D crystals. It is not necessary to aim for a homogeneous appearance of membranes, as the largest and most well-ordered 2D crystals are selected visually for data collection. These types of samples will be easily recognized when the crystallization trials are repeated as well as when the samples are used for cryo-EM data collection, resulting in a maximum number of high resolution images.
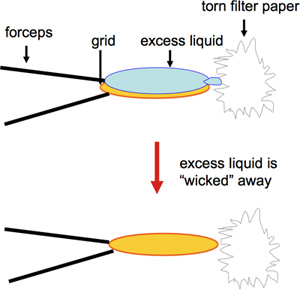
Figure 1. This figure shows blotting the edge of the grid with a torn piece of Whatman #4 filter paper.
Discussion
Proper evaluation of samples requires careful assessment of a sufficient number of membranes. For example, samples with as low as 2% crystalline arrays out of over 180 imaged proteoliposomes gave critical information for quick optimization of 2D crystallization conditions 7.
When precipitation of the protein occurs, a grid might be abandoned from further screening after inspection at low magnification, although occasional partial precipitation of protein occurs. Even very small m...
Acknowledgements
We thank our collaborators for providing valuable protein samples, which contributed to some of our methods related experience and observations. Günther Schmalzing kindly provided the opportunity to FR to join this project. Barbara Armbruster, Jacob Brink and Deryck Mills are thanked for their outstanding help and input on equipment. Funding was provided by NIH grant HL090630.
Materials
| Name | Company | Catalog Number | Comments |
| 400-mesh copper TEM grids coated with carbon film | |||
| forceps: regular and anti-capillary | Dumont #5 and Dumont N5AC or similar | ||
| Micropipette and pipette tips | |||
| Whatman #4 filter paper | |||
| 1% uranyl acetate | |||
| Dialysis sample to be screened for 2D crystals | |||
| Glycerol/sucrose-free dialysis buffer | Optional | ||
| JEOL-1400 transmission electron microscope (TEM) | similar 80 – 120kV TEM equipped with an Lab6 or tungsten filament and film and/or CCD cameras (Gatan Orius SC1000 and/or UltraScan1000 CCD cameras and Gatan Digitial Micrograph software package or Tietz cameras (TVIPS)) |
References
- Johansen, B. V. Bright field electron microscopy of biological specimens V. A low dose pre-irradiation procedure reducing beam damage. Micron. 7, 145-156 (1976).
- Schmidt-Krey, I. Electron crystallography of membrane proteins: Two-dimensional crystallization and screening by electron microscopy. Methods. 41, 417-426 (2007).
- Wang, D. N., Kühlbrandt, W. High-resolution electron crystallography of light-harvesting chlorophyll a/b-protein complex in three different media. J Mol Biol. 217, 691-699 (1991).
- Schmidt-Krey, I., Rubinstein, J. L. Electron cryomicroscopy of membrane proteins: specimen preparation for two-dimensional crystals and single particles. Micron. , (2010).
- Schmidt-Krey, I., Mutucumarana, V., Haase, W., Stafford, D. W., Kühlbrandt, W. Two-dimensional crystallization of human vitamin K-dependent γ-glutamyl carboxylase. J Struct Biol. 157, 437-442 (2007).
- Trachtenberg, S., DeRosier, D. J., Zemlin, F., Beckmann, E. Non-helical perturbations of the flagellar filament: Salmonella typhimurium SJW117 at 9.6 Å resolution. J Mol Biol. 276, 759-773 (1998).
- Zhao, G., Johnson, M. C., Schnell, J. R., Kanaoka, Y., Irikura, D., Lam, B. K., Austen, K. F., Schmidt-Krey, I. Two-dimensional crystallization conditions of human leukotriene C4 synthase requiring a particularly large combination of specific parameters. J Struct Biol. 169, 450-454 (2010).
- Cheng, A., Leung, A., Fellmann, D., Quispe, J., Suloway, C., Pulokas, J., Abeyrathne, P. D., Lam, J. S., Carragher, B., Potter, C. S. Towards automated screening of two-dimensional crystals. J Struct Biol. 160, 324-331 (2007).
- Vink, M., Derr, K. D., Love, J., Stokes, D. L., Ubarretxena-Belandia, I. A high-throughput strategy to screen 2D crystallization trials of membrane proteins. J Struct Biol. 160, 295-304 (2007).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved