A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Synthesis of Cyclic Polymers and Characterization of Their Diffusive Motion in the Melt State at the Single Molecule Level
In This Article
Summary
A protocol for the synthesis and characterization of diffusive motion of cyclic polymers at the single molecule level is presented.
Abstract
We demonstrate a method for the synthesis of cyclic polymers and a protocol for characterizing their diffusive motion in a melt state at the single molecule level. An electrostatic self-assembly and covalent fixation (ESA-CF) process is used for the synthesis of the cyclic poly(tetrahydrofuran) (poly(THF)). The diffusive motion of individual cyclic polymer chains in a melt state is visualized using single molecule fluorescence imaging by incorporating a fluorophore unit in the cyclic chains. The diffusive motion of the chains is quantitatively characterized by means of a combination of mean-squared displacement (MSD) analysis and a cumulative distribution function (CDF) analysis. The cyclic polymer exhibits multiple-mode diffusion which is distinct from its linear counterpart. The results demonstrate that the diffusional heterogeneity of polymers that is often hidden behind ensemble averaging can be revealed by the efficient synthesis of the cyclic polymers using the ESA-CF process and the quantitative analysis of the diffusive motion at the single molecule level using the MSD and CDF analyses.
Introduction
Cyclic polymers are unique in that they do not have chain ends. They often exhibit unusual behaviors which is distinct from their linear counterpart, including increased thermal stability of polymer micelles by a linear-to-cyclic conversion,1,2 and spatial organization of DNA in bacterial cells by a loop formation.3 Topological interactions between the cyclic chains are believed to be the critical factor for such unusual behaviors.4,5 Therefore, characterizing the motion and relaxation of cyclic polymers under entangled conditions has been an important research topic in polymer science for decades.6
Cyclic polymer dynamics has been investigated using both synthetic and naturally occurring molecules by means of ensemble averaged experimental methods such as nuclear magnetic resonance (NMR), light scattering, and viscosity measurements.7-9 However, these studies often suffer from impurity molecules in the samples.10 Furthermore, spatiotemporal heterogeneities of the motion of individual molecules caused by inherent structural heterogeneity of entangled polymers are often hidden behind the ensemble averaging in these studies. In order to characterize molecular level dynamics of cyclic polymers, a synthesis method that provides high purity cyclic polymers and an experimental and analysis methods that allow for quantitative characterization of molecular motion at the single molecule level have to be developed. Here, we will show a method to synthesize high-purity cyclic and dicyclic poly(THF)s that incorporate a fluorophore unit using an electrostatic self-assembly and covalent fixation (ESA-CF) process11-13 and a method to analyze the motion of the individual fluorophore-incorporated polymer chains using a combination of mean-squared displacement (MSD) and cumulative distribution function (CDF) analyses.
A proper data processing has been shown to be essential for the accurate characterization of the diffusive motion. With an adequate MSD and CDF analyses, a multiple-mode diffusion of the cyclic and dicyclic polymers in the melt and semi-dilute solution of the linear polymer chains has been revealed,14-16 suggesting the significant effects of the topological states of the polymers on the diffusive motion of the chains under entangled conditions.17 While the experimental and analytical approaches to characterize the motion of the cyclic polymers are described in this protocol, the same method can be used to quantitatively characterize the diffusive motion in many other heterogeneous systems. The approach would be especially suitable when multiple diffusion components existing in the samples are to be analyzed.
Protocol
1. Synthesis of Monofunctional and Bifunctional Poly(THF)
- Monofunctional poly(THF)
- Flame dry a 2-neck 100-ml round-bottom flask. Vacuum and fill the flask with nitrogen (3 cycles).
- Add distilled tetrahydrofuran (THF) (50 ml) to the flask. Put the flask in a water bath at 20 °C and equilibrate the temperature.
- Add methyl triflate (0.5 mmol) to the flask by a syringe. Stir the mixture for 5-10 min at 20 °C.
- Add N-phenyl pyrrolidine (4-6 equiv.) to the flask by a syringe. Stir the mixture for 30-60 min.
- Completely remove the solvent under reduced pressure (ca. 100 Torr). Dissolve the residue in 3-5 ml of acetone. Add the acetone solution into 300-500 ml of n-hexane. Filter the precipitate and dry it under reduced pressure.
- Bifunctional poly(THF)
- Flame dry a 2-neck 100-ml round-bottom flask. Vacuum and fill the flask with nitrogen (3 cycles).
- Add distilled THF (50 ml) to the flask. Put the flask in a water bath at 20 °C and equilibrate the temperature.
- Add triflic anhydride (0.3 mmol) to the flask by a syringe. Stir the mixture for 5-10 min at 20 °C.
- Add N-phenyl pyrrolidine (4-6 equiv.) to the flask by a syringe. Stir the mixture for 30-60 min.
- Completely remove the solvent under reduced pressure (ca. 100 Torr). Dissolve the residue in 3-5 ml of acetone. Add the acetone solution to 300-500 ml of n-hexane. Filter the precipitate and dry it under reduced pressure.
2. Synthesis of Perylene Diimide-incorporated 4-armed Star and 8-shaped Dicyclic Poly(THF)
- Armed star poly(THF)
- Ion exchange
- Dissolve perylene diimide tetracarboxylate sodium salt in water (10 mg/ml, 150 ml). Dissolve monofunctional poly(THF) in acetone (160 mg/ml, 4 ml). Add the acetone solution dropwise into the vigorously stirred aqueous solution. Collect the formed precipitate by filtration.
- Repeat the above procedure with the recovered precipitate (2.1.1.1) four times.
- Covalent fixation
- Dissolve the obtained precipitate in toluene (5 mg/ml). Reflux the solution for 4 hr.
- Completely remove the solvent under reduced pressure (ca. 100 Torr). Filter the residue through a plug of silica gel with n-hexane/acetone (2/1 vol/vol). Add the solution into ice-cooled water (300-500 ml) to precipitate the product. Collect the precipitate by filtration.
- Ion exchange
- Dicyclic 8-shaped poly(THF)
- Ion exchange
- Dissolve perylene diimide tetracarboxylate sodium salt in water (6 mg/ml, 50 ml). Dissolve bifunctional poly(THF) (0.5 g) in 30-50 ml of acetone. Add the acetone solution dropwise into the vigorously stirred aqueous solution at 0 °C. Collect the formed precipitate by filtration.
- Repeat the above procedure with the recovered precipitate (2.2.1.1).
- Covalent fixation
- Dissolve the obtained precipitate in toluene (0.05 g/L). Reflux the solution for 4 hr.
- Completely remove the solvent under reduced pressure (ca. 100 Torr). Add toluene to partially dissolve the residue. Re-precipitate into 300-500 ml of n-hexane.
- Filter the formed precipitate through a plug of silica gel with n-hexane/acetone (2/1 vol/vol). Re-precipitate into 300-500 ml of water.
- Purify the formed precipitate by column chromatography18 using a polystyrene gel. Further purify the crude product by preparative gel permeation chromatography (GPC)19 with an eluent of CHCl3 to remove byproducts by monitoring refractive index (RI) and UV detectors.
- Ion exchange
3. Single-molecule Fluorescence Imaging Experiment
- Sample preparation
- Cleaning of microscope cover slips
- Place No. 1.5 24 x 24 mm microscope cover slips in a staining jar.
- Add 1 M potassium hydride solution (100 ml) into the jar and sonicate for 15 min. Pour off the potassium hydroxide solution by decantation and rinse the cover slips with ultra-pure water for several times. Add spectroscopic grade ethanol (100 ml) into the jar and sonicate for 15 min.
- Pour off the ethanol by decantation and rinse the cover slips with ultra-pure water for several times. After pouring off the ultra-pure water by decantation, repeat the step 3.1.1.2.
- Add ultra-pure water to the jar and sonicate for 15 min. Rinse the cover slips with ultra-pure water for several times. Take out the cover slips from the jar by a plastic tweezers and dry them by either dry air or dry nitrogen.
- Preparation of polymer melt samples14,15
- Add 100 µl of non-labeled linear poly(THF) in a glass bottle and heat it to a temperature above the melting point (approximately 25 °C) using a hair dryer.
- Dissolve the fluorophore-incorporated polymer (linear, 4-armed star, cyclic, or 8-shaped dicyclic synthesized in 2.1 and 2.2) in chloroform (1 ml, 10-6 M). Add 1 µl of the solution to the 100 µl of the melt of the non-labeled linear poly(THF).
- After thoroughly mixing the sample with a pipette tip, evaporate chloroform by heating the sample using a dryer.
NOTE: This provides a melt of the non-labeled linear poly(THF) containing 10-8 M of the fluorophore incorporated polymers. - Take 10 µl of the sample using a micro-pipette and drop it on a cleaned cover slip. Put another cleaned cover slip on the sample and sandwich the sample between the two cover slips.
- Press the sample gently using a plastic tweezers.
- Cleaning of microscope cover slips
- Wide-field fluorescence imaging setup15
- Introduction of an excitation laser (488 nm) into the back port of the inverted microscope
- Insert an excitation bandpass filter and polarizer into the beam path.
- Expand the beam to approximately 1 cm in diameter by a beam expander.
- Insert a quarter wave plate into the beam path. Set the optical axis of the wave plate at 45 degree with respect to that of the polarizer. Alternatively, insert a Berek compensator and set the optical delay to λ/4.
- Insert a diaphragm in the excitation beam path to adjust the beam size.
- Before introducing the laser beam into the back port of the inverted optical microscope, insert a focusing lens (plan-convex lens, focal length ≈ 300 mm) at a position where the laser beam out of the objective lens is collimated.
- After reflecting the laser beam using a dichroic mirror mounted on a filter cube, introduce the laser beam to the sample through a high numerical aperture (NA) objective lens (e.g., NA 1.3, 100X magnification, oil immersion).
- Attach an objective heater to the objective lens and set the temperature to 30 °C.
- Mounting the sample on the stage of the inverted microscope
- Drop one drop of immersion oil on the objective lens and mount the sample on the microscope state.
- Ensure that the sample thickness of approximately 10 µm is obtained by checking the axial position of the bottom and top surface of the sample.
- Adjust the focus of the microscope to a few micrometers above the bottom surface of the sample.
- Obtain circularly polarized excitation light under the objective lens
- Insert a polarizer into the collimated laser beam out of the objective lens.
- Record the intensity of the laser transmitted through the polarizer by inserting a power meter after the polarizer. Record the transmitted laser power at different polarization angles by rotating the polarizer.
- If the transmitted laser power is not constant at all polarization angles, slightly rotate the quarter wave plate or Berek compensator inserted in the excitation beam path.
- Repeat step 3.2.5.2 and 3.2.5.3 until the constant transmitted laser power is obtained at all polarization angles. Ensure that the circularly polarized light is obtained at the sample.
- Setup the EM (electron multiplying)-charge coupled device (CCD) camera
- Attach the EM-CCD camera to the side port of the microscope and connect it to the image acquisition software.
- If necessary, synchronize the camera exposure to a mechanical shutter or acousto-optical tunable filter inserted in the excitation beam path by sending the transistor-transistor logic (TTL) signals generated by the EM-CCD camera to the devices. Alternatively, synchronize the camera exposure to the laser output by sending the TTL signals generated by the EM-CCD camera to the laser.
NOTE: The latter option is applicable only when a solid-state laser whose output power can be modulated by input transistor-transistor logic (TTL) signals is used for the experiment. - Apply an EM gain (typically approximately 300) to the CCD camera using software controlling the camera in order to obtain a high quality fluorescence image of the single fluorophore.
- Set a region of interest (ROI) (typically 128 x 128 pixels at the center of the field of view) using software controlling the camera.
NOTE: This allows for the imaging experiments at the frame rates of 100 - 200 Hz in the frame transfer mode, which is required for visualizing the motion of the fluorophore-incorporated polymer chains in the melt sample.
- Introduction of an excitation laser (488 nm) into the back port of the inverted microscope
- Running the experiment
- Optimizing the experimental conditions
- Adjust the illumination area of the sample to approximately 20 µm in diameter using the diaphragm inserted in the excitation beam path.
- Set the excitation laser power at the sample to 4 - 8 mW by manually selecting an appropriate neutral density (ND) filter inserted in the excitation beam path.
NOTE: This provides the mean laser power of 1 - 2 kW cm-2 at the sample. - Record fluorescence images of the sample at the frame rates of 100 - 200 Hz. If the fluorescence intensity obtained from the individual fluorophore-incorporated polymers is too low, gradually increase the excitation power using the ND filter until reaching approximately 100 mW at the sample.
- If the quality of the single-molecule fluorescence image is still not satisfactory, check the fluorescence impurities in the sample by recording fluorescence images of a pure melt of the non-labeled poly(THF). In case a high fluorescence background is observed, use different non-labeled poly(THF).
- If the density of the fluorescence spot obtained from the fluorophore-incorporated polymers in the melt is too high to spatially isolate them (this causes errors in the analysis of the diffusive motion), decrease the concentration of the fluorophore-incorporated polymers in the sample until spatially isolated spots are observed.
- If the density of the fluorescence spot obtained from the fluorophore-incorporated polymers in the melt is too low (this causes a low throughput of the imaging experiment), increase the concentration of the fluorophore-incorporated polymers in the sample until an appropriate density of the fluorescence spot is reached.
- If the fluorescence images obtained from the fluorophore-incorporated polymers in the melt are blurred, increase the frame rates of the imaging acquisition.
NOTE: This often requires a smaller ROI, typically 64 x 64 pixels.
- Optimizing the experimental conditions
- Image acquisition
- Once the experimental conditions are optimized, leave the mounted sample on the microscope stage for an hour so that the sample reaches equilibrium conditions.
- Record 500 - 1,000 fluorescence image sequences of the fluorophore-incorporated polymers in the melt state at a 100 - 200 Hz frame rate. If the default file format is not TIFF, convert all the image sequences to the TIFF format.
4. Analysis of the Diffusive Motion
- Mean-squared displacement (MSD) analysis
- Crop the fluorescence image sequences in such a way that each image sequence contains a single and well-focused diffusing fluorophore-incorporated polymer using image processing software, such as ImageJ.
- When the cropped image sequences contain more than 10 frames, split the image sequences into multiple sequences such that each sequence consists of 10 frames.
- Determine the positions of the molecules in each image sequences accurately by two-dimensional Gaussian fitting of the images.
- Determine the diffusion coefficient (D) of individual molecules by mean-squared displacement (MSD) analysis of the diffusion trajectories (i.e., time-dependent positions of the molecule) using an equation20
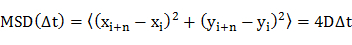
where xi and yi are the positions of the molecule in the image frame i, and n denotes the frame number with the time lapse Δt from frame i. - Plot the diffusion coefficients in a frequency histogram.
NOTE: Typically, the histogram is constructed from more than 100 molecules.
- Cumulative distribution function (CDF) analysis
NOTE: A CDF, P(r2, iΔt) corresponds to the cumulative probability of finding the diffusing molecules within a radius r from the origin after a certain time lag iΔt.- Calculate the squared-displacement occurring during time lags of 1Δt, 2Δt, ····, iΔt for all the diffusion trajectories obtained in 4.1.3.
NOTE: These calculations give total mi squared-displacements for the time lags of iΔt. - Calculate numbers of the squared-displacements (li) within total mi data set that are smaller than r2 at different r2 values (0 < r2 < ∞). Normalized li vs r2 plots correspond to the CDF, P(r2, iΔt).
- Calculate the squared-displacement occurring during time lags of 1Δt, 2Δt, ····, iΔt for all the diffusion trajectories obtained in 4.1.3.
- Analysis of CDFs with distinct diffusion models
Note: The obtained CDFs are fitted by distinct diffusion models; homogeneous diffusion model, multiple diffusion modes in which the D distribution is described by a Gaussian (single Gaussian model), and multiple diffusion modes in which the D distribution is described by multiple Gaussian (multiple Gaussian model).- In the homogeneous diffusion model, determine a mean D by fitting the CDF using an equation21
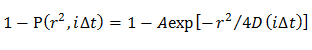
NOTE: Any deviation from the equation suggests the heterogeneous diffusion of the molecule. - In the single Gaussian model, determine the probability distribution of D described by a Gaussian (f(D)) by fitting the CDF using15
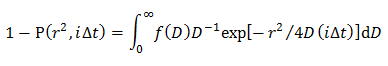
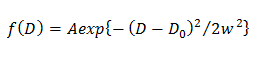
where A, w, and D0 are the amplitude, width, and center of the Gaussian. - In the double Gaussian model, determine the probability distribution of jth component of D described by a Gaussian (f(D)) by fitting the CDF using14
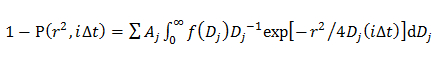

where Aj is the fraction of each diffusion component, and αj, wj, and D0j are the amplitude, width, and center of the jth component of the Gaussian.
- In the homogeneous diffusion model, determine a mean D by fitting the CDF using an equation21
- Calculation of the theoretical probability distribution of diffusion coefficient
NOTE: The probability distributions of D occurring due to the statistical errors (p(D)dD) are calculated for the different diffusion models; homogeneous diffusion model, multiple diffusion modes in which the D distribution is described by a Gaussian (single Gaussian model), and multiple diffusion modes in which the D distribution is described by multiple Gaussian (multiple Gaussian model).- In the homogeneous diffusion model, calculate the statistical probability distribution of D using an equation22
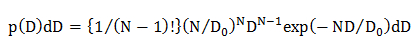
where N is the number of the data points in a diffusion trajectory (N = 10, see 4.1.2), D0 is the mean diffusion coefficient (determined by the CDF analysis, see 4.2.3.1), and D is the experimentally obtained diffusion coefficient for an individual trajectory. - In the single Gaussian diffusion model, calculate the statistical probability distribution of D using an equation15

where f(D) denotes probability distribution of D determined by the CDF analysis (see 4.2.3.2), and D0 is the mean diffusion coefficient (determined by the CDF analysis, see 4.2.3.2). - In the double Gaussian diffusion model, calculate the statistical probability distribution of D using an equation14

where f(Dj) denotes probability distribution of the jth component of D (Dj) determined by the CDF analysis (see 4.2.3.3), and D0j is the mean diffusion coefficient of the jth component (determined by the CDF analysis, see 4.2.3.3).
- In the homogeneous diffusion model, calculate the statistical probability distribution of D using an equation22
Results
The perylene diimide-incorporated 4-armed star and 8-shaped dicyclic poly(THF)s were synthesized using the electrostatic self-assembly and covalent fixation (ESA-CF) process (Figure 1, Figure 2). Time-lapse single-molecule fluorescence images were measured for the 4-armed (Figure 3a) and 8-shaped (Figure 3b) polymers. The time-lapse fluorescence images (Figure 3) show spatially isolated bright and sharp s...
Discussion
The 4-armed and 8-shaped polymers were prepared via the ESA-CF protocol (Figure 1), which is a critical step for the synthesis.12,24 Monofunctional and bifunctional linear poly(THF)s with N-phenylpiperidinium end groups were synthesized according to the previous procedure.11 The ion exchange was carried out by reprecipitation of an acetone solution of a polymer precursor with triflate counteranions into an aqueous solution containing an excess amount of carboxylate.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research No. 22750122 (S.H.), No. 26288099 (T.Y.), and No. 23350050 (Y.T.) of the Japan Society for the Promotion of Science. S.H. is grateful for The Kurata Memorial Hitachi Science and Technology Foundation. The research reported in this publication was supported by the King Abdullah University of Science and Technology (S.H.).
Materials
| Name | Company | Catalog Number | Comments |
| Materials | |||
| THF | Godo | ||
| Wakosil C-300 | Wako Pure Chemical Industries | ||
| Acetone | Godo | ||
| Toluene | Godo | ||
| n-Hexane | Godo | ||
| CHCl3 | Kanto Chemical | ||
| Bio-Beads S-X1 | Bio-Rad | ||
| Methyl triflate | Nacalai Tesque | ||
| Triflic anhydride | Nacalai Tesque | ||
| Potassium Hydroxide | Wako Pure Chemical Industries | ||
| Ethanol | Wako Pure Chemical Industries | ||
| Poly(tetrahydrofuran) | Aldrich | ||
| Chloroform | Wako Pure Chemical Industries | ||
| Immersion oil | Cargille | Type 37 / Type A | |
| Equipment | |||
| 2-Neck 100-ml round-bottom flask | |||
| Flask | |||
| Beaker | |||
| Funnel | |||
| Filter paper | Whatman | ||
| Reflux condenser | |||
| Syringe | |||
| Water bath | |||
| Magnetic stirrer | |||
| Rotary evaporator | |||
| Microscope cover slips (24 x 24 mm, No. 1) | Matsunami Glass | CO22241 | |
| Staining jar | AS ONE Corporation | 1-7934-01 | |
| Ultrasonic cleaner | VWR International | 142-0047 | |
| Inverted microscope | Olympus | IX71 | |
| Ar-Kr ion laser | Coherent | Innova 70C | |
| Berek compensator | Newport | 5540 | |
| Excitation filter | Semrock | LL01-488-12.5 | |
| Dichloric mirror | Omega optical | 500DRLP | |
| Emission filter | Semrock | BLP01-488R-25 | |
| Lens and mirror | Thorlabs | ||
| EM-CCD camera | Andor Technology | iXon | |
| Objective lens (100X, N.A. = 1.3) | Olympus | UPLFLN 100XOP | |
| Objective heater | Bioptechs | ||
| Preparative GPC | Japan Analytical Industry | LC-908 |
References
- Honda, S., Yamamoto, T., Tezuka, Y. Topology-Directed Control on Thermal Stability: Micelles Formed from Linear and Cyclized Amphiphilic Block Copolymers. J. Am. Chem. Soc. 132 (30), 10251-10253 (2010).
- Honda, S., Yamamoto, T., Tezuka, Y. Tuneable enhancement of the salt and thermal stability of polymeric micelles by cyclized amphiphiles. Nat. Commun. 4, (2013).
- Jun, S., Mulder, B. Entropy-driven spatial organization of highly confined polymers: Lessons for the bacterial chromosome. Proc. Natl. Acad. Sci. U. S. A. 103 (33), 12388-12393 (2006).
- McLeish, T. Polymers without beginning or end. Science. 297 (5589), 2005-2006 (2002).
- McLeish, T. Polymer dynamics: Floored by the rings. Nat. Mater. 7 (12), 933-935 (2008).
- Roovers, J., Tezuka, Y. . Topological Polymer Chemistry: Progress of Cyclic Polymers in Syntheses, Properties and Functions. , 137-156 (2013).
- Klein, J. Evidence for reptation in an entangled polymer melt. Nature. 271 (5641), 143-145 (1978).
- Leger, L., Hervet, H., Rondelez, F. Reptation in entangled polymer-solutions by forced rayleigh light-scattering. Macromolecules. 14 (6), 1732-1738 (1981).
- von Meerwall, E. D., Amis, E. J., Ferry, J. D. Self-diffusion in solutions of polystyrene in tetrahydrofuran - comparison of concentration dependences of the diffusion-coefficients of polymers, and a ternary probe component. Macromolecules. 18 (2), 260-266 (1985).
- Kapnistos, M., et al. Unexpected power-law stress relaxation of entangled ring polymers. Nat. Mater. 7 (12), 997-1002 (2008).
- Adachi, K., Takasugi, H., Tezuka, Y. Telechelics having unstrained cyclic ammonium salt groups for electrostatic polymer self-assembly and ring-emitting covalent fixation. Macromolecules. 39 (17), 5585-5588 (2006).
- Oike, H., Imaizumi, H., Mouri, T., Yoshioka, Y., Uchibori, A., Tezuka, Y. Designing unusual polymer topologies by electrostatic self-assembly and covalent fixation. J. Am. Chem. Soc. 122 (40), 9592-9599 (2000).
- Yamamoto, T., Tezuka, Y. Topological polymer chemistry: a cyclic approach toward novel polymer properties and functions. Polym. Chem. 2 (9), 1930-1941 (2011).
- Habuchi, S., Fujiwara, S., Yamamoto, T., Tezuka, Y. Single-molecule imaging reveals topological isomer-dependent diffusion by 4-armed star and dicyclic 8-shaped polymers. Polym. Chem. 6 (22), 4109-4115 (2015).
- Habuchi, S., Fujiwara, S., Yamamoto, T., Vacha, M., Tezuka, Y. Single-Molecule Study on Polymer Diffusion in a Melt State: Effect of Chain Topology. Anal. Chem. 85 (15), 7369-7376 (2013).
- Habuchi, S., Satoh, N., Yamamoto, T., Tezuka, Y., Vacha, M. Multimode Diffusion of Ring Polymer Molecules Revealed by a Single-Molecule Study. Angew. Chem. Int. Ed. 49 (8), 1418-1421 (2010).
- Habuchi, S., Tezuka, Y. . Topological Polymer Chemistry: Progress of Cyclic Polymers in Syntheses, Properties and Functions. , 265-290 (2013).
- Fernandez, P., Bayona, J. M. Use of off-line gel-remeation chromatography normal-phase liquid-chromatography fro the determination of polycyclic aromatic-compounds in environmental-samples and standard reference materials (air particulate matter and marine sediment). J. Chromatogr. 625 (2), 141-149 (1992).
- Biesenberger, J. A., Tan, M., Duvdevan, I., Maurer, T. Recycle gel permeation chromatography. 1. recycle principle and design. J. Polym. Sci. Pol. Lett. 9 (5), 353 (1971).
- Kusumi, A., Sako, Y., Yamamoto, M. Confined lateral diffusion of membrane-receptors as studied by single-particle tracking (nanovid microscopy) - effects of calcium-induced differentiation in cultured epithelial-cells. Biophys. J. 65 (5), 2021-2040 (1993).
- Schutz, G. J., Schindler, H., Schmidt, T. Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys. J. 73 (2), 1073-1080 (1997).
- Vrljic, M., Nishimura, S. Y., Brasselet, S., Moerner, W. E., McConnell, H. M. Translational diffusion of individual class II MHC membrane proteins in cells. Biophys. J. 83 (5), 2681-2692 (2002).
- Margineanu, A., et al. Photophysics of a water-soluble rylene dye: Comparison with other fluorescent molecules for biological applications. J. Phys. Chem. B. 108 (32), 12242-12251 (2004).
- Tezuka, Y., Oike, H. Self-assembly and covalent fixation for topological polymer chemistry. Macromol. Rapid Commun. 22 (13), 1017-1029 (2001).
- Deres, A., et al. The Origin of Heterogeneity of Polymer Dynamics near the Glass Temperature As Probed by Defocused Imaging. Macromolecules. 44 (24), 9703-9709 (2011).
- Flier, B. M. I., et al. Heterogeneous Diffusion in Thin Polymer Films As Observed by High-Temperature Single-Molecule Fluorescence Microscopy. J. Am. Chem. Soc. 134 (1), 480-488 (2012).
- Habuchi, S., Oba, T., Vacha, M. Multi-beam single-molecule defocused fluorescence imaging reveals local anisotropic nature of polymer thin films. Phys. Chem. Chem. Phys. 13 (15), 6970-6976 (2011).
- Zettl, U., et al. Self-Diffusion and Cooperative Diffusion in Semidilute Polymer Solutions As Measured by Fluorescence Correlation Spectroscopy. Macromolecules. 42 (24), 9537-9547 (2009).
- Kirstein, J., Platschek, B., Jung, C., Brown, R., Bein, T., Brauchle, C. Exploration of nanostructured channel systems with single-molecule probes. Nat. Mater. 6 (4), 303-310 (2007).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved