A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Identification of Fatty Acids in Bacillus cereus
In This Article
Summary
We propose a protocol to identify fatty acids without the need to purify them. It combines information on the retention times with the mass spectra of three types of fatty acid derivatives: fatty acid methyl esters (FAMEs), 4,4-dimethyl oxazoline derivatives (DMOX), and 3-pyridylcarbinyl esters (picolinyl).
Abstract
The Bacillus species contain branched chain and unsaturated fatty acids (FAs) with diverse positions of the methyl branch (iso or anteiso) and of the double bond. Changes in FA composition play a crucial role in the adaptation of bacteria to their environment. These modifications entail a change in the ratio of iso versus anteiso branched FAs, and in the proportion of unsaturated FAs relative to saturated FAs, with double bonds created at specific positions. Precise identification of the FA profile is necessary to understand the adaptation mechanisms of Bacillus species.
Many of the FAs from Bacillus are not commercially available. The strategy proposed herein identifies FAs by combining information on the retention time (by calculation of the equivalent chain length (ECL)) with the mass spectra of three types of FA derivatives: fatty acid methyl esters (FAMEs), 4,4-dimethyl oxazoline derivatives (DMOX), and 3-pyridylcarbinyl ester (picolinyl). This method can identify the FAs without the need to purify the unknown FAs.
Comparing chromatographic profiles of FAME prepared from Bacillus cereus with a commercial mixture of standards allows for the identification of straight-chain saturated FAs, the calculation of the ECL, and hypotheses on the identity of the other FAs. FAMEs of branched saturated FAs, iso or anteiso, display a constant negative shift in the ECL, compared to linear saturated FAs with the same number of carbons. FAMEs of unsaturated FAs can be detected by the mass of their molecular ions, and result in a positive shift in the ECL compared to the corresponding saturated FAs.
The branching position of FAs and the double bond position of unsaturated FAs can be identified by the electron ionization mass spectra of picolinyl and DMOX derivatives, respectively. This approach identifies all the unknown saturated branched FAs, unsaturated straight-chain FAs and unsaturated branched FAs from the B. cereus extract.
Introduction
Fatty acid methyl ester (FAME) gas chromatography (GC) is an essential method for lipid characterization. It rapidly separates and quantifies the various fatty acids (FAs) of a sample after a short extraction step. Derivatives of methyl esters are highly volatile, stable and inert toward the chromatographic column, thereby avoiding tailing peaks. Their identification is rather straightforward when the sample consists of well-known FAs because the chromatographic profiles are either published or compared to standards. In addition, the repeated injection of calibration standards for quantification of various FAs is not required, given their almost constant response to flame ionization detection (FID)1.
In addition to FID, mass spectrometry (MS) detection provides a complementary set of information to confirm FAMEs. However, when FAMEs are charged using electron ionization (EI), the resulting spectra do not always allow for the identification of FA fine structure. For instance, branching position (i.e., a branched methyl group) is difficult to predict because the diagnostic ions are difficult to detect1 and the characteristic change in target ion abundance is machine-dependent, preventing the use of mass spectra libraries2. Another challenge lies in identifying the double bond position because EI causes double bond migration. Thus, FA isomers with varying double bond positions cannot be differentiated by their mass spectra. Fortunately, other tools have been developed for FA identification. For instance, the presence and the position of branching or of double bonds in FAs can be conjectured by calculating the equivalent chain length (ECL)3.
Other derivatization methods result in different mass spectra, dependent on the location of a double bond or a branched methyl group. 4,4-Dimethyl oxazoline derivatives (DMOX)4 allow for easy identification of the position of monounsaturated fatty acid double bonds. 3-pyridylcarbinyl ester (picolinyl ester) derivatives allow for the unambiguous identification of the location of methyl branched FAs5. Combining chromatographic retention (ECL) and mass spectra (DMOX and picolinyl) information allows for the identification of most FAs without the need to use complex methods of purification, as required for nuclear magnetic resonance (NMR) spectrum, the uncontestable method for structural characterization1.
Bacteria of the genus Bacillus, which include some human and animal pathogens, are able to colonize highly diverse niches and are therefore widely distributed in the environment6. Among the Bacillus genus, FA composition is influenced by the ecological niche of the species with modulations in FA patterns to adapt to a wide range of environmental changes (e.g., growth medium, temperature, pH, etc.)7-9. Because of the relative homogeneity of the FA pattern across species of the genus Bacillus during growth in standardized conditions, determination of FA composition is one of the essential criteria used to define the Bacillus species. A unique attribute of the Bacillus genus is the abundance of branched-chain FAs containing 12-17 carbons10-12 with the ratio between iso and anteiso isomers being a key determinant of adaptation to environmental conditions. Bacillus species also adapt to environmental fluctuations by altering the proportion of unsaturated fatty acids. In some species, such as Bacillus cereus, two fatty acid desaturases create double bonds in different positions of the alkyl chain13 with different roles in adaptation9. The example of the Bacillus genus illustrates the importance of precisely identifying the double bond position and FA branching. Collectively, identification of Bacillus FA patterns has several useful applications. Herein, we propose a novel GC-MS approach for Bacillus FA pattern identification that overcomes the inherent limitations of a classical GC-MS analysis.
This innovative approach can be used directly on raw biological material, and consists of a combination of existing techniques: information on retention times (ECL) and mass spectra of different FAs derivatives (FAME, DMOX and picolinyl-ester).
We use the following FA nomenclature. i, a, and n indicate iso, anteiso methyl branched, and straight-chain fatty acid, respectively. Unsaturated FAs were named by C:d where C is the number of carbon atoms in the fatty acid and d is the number of double bonds. Δx indicates the position of the double bond, where the double bond is located on the xth carbon-carbon bond, counting from the carboxylic acid end.
Protocol
1. Bacterial Cultures
- Prepare a lawn of the bacteria (Bacillus cereus strain ATCC 14579) by spreading 100 µl of an overnight culture of the strain incubated at 30 °C in LB (Luria-Bertani medium), over the surface of a plate of LB agar medium. Incubate the plate overnight at 30 °C.
2. ECL: Equivalent Chain Length
- Calculate ECL as follows:
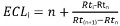 with:
with:
i, the solute of interest;
n, the carbon number of the straight chain saturated fatty acid methyl ester eluting before solute I;
n+1, the carbon number of the straight chain saturated fatty acid methyl ester eluting after solute I;
Rti, Rtn, Rt(n+1) the retention times of the FAME peaks described above.
NOTE: Obtain the retention times of straight chain saturated fatty acids methyl esters by injection of a mixture of standards (BAME).
3. FAME Preparation and Analysis
- In order to obtain lipid fatty acids, harvest bacterial cells by scraping colonies from the agar plate and transfer 40 mg (fresh weight, equivalent to 109 viable cells) of bacteria into a 10 ml glass tube with screw caps and PTFE seals.
- Perform transesterification via the ester link method14,15 detailed below.
- Add 5 ml of 0.2 M KOH in methanol to the fresh bacterial cells and incubate at 37 °C for 1 hr. This reaction consists of alkaline methanolysis, breaking the ester link in the lipid and producing fatty acid methyl esters.
- Add 1 ml of 1 M acetic acid to lower the pH to 7.0. Check the pH with pH test strips.
- Add 3 ml of hexane to extract FAMEs.
- Transfer supernatant (organic phase) into clean tubes and concentrate by evaporation at room temperature under a continuous flow of nitrogen to obtain approximately 200 µl of extract. Transfer the sample into a GC vial with insert.
- Inject extracts into a gas chromatography-mass spectrometry (GC-MS) system.
4. GC/MS Conditions
- Inject FAME samples into a GC-MS instrument equipped with a capillary column ZB-WAX (length, 30 m; diameter, 0.25 mm; film thickness, 0.25 µm).
- Set the injection port (in splitless mode) temperature to 250 °C. Use helium as a carrier gas, with a linear velocity of 37 cm/sec. Hold the oven temperature at 50 °C for 1 min, increase to 190 °C at a rate of 20 °C/min, and increase further to a final temperature of 230 °C at a rate of 2 °C/min.
- For the MS, record the mass spectra by electron ionization (EI) at 70 eV, and set the acquisition of the total ion current between 50 and 400 atomic mass units (amu) (2 scans/sec).
- When required, inject DMOX and picolinyl derivatives under the same condition except the temperature program oven as follows:
DMOX: 50 °C (1 min), 20 °C/min until 210 °C and 2 °C/min until 240 °C (5 min);
Picolinyl: 6 °C (1 min), 20 °C/min until 220 °C and 2 °C/min until 250 °C (20 min).
5. Picolinyl Ester Preparation from FAME16
- Evaporate the FAME extract from Section 3 with a flow of nitrogen (at least 10 mg dry material) and dissolve in 1 ml of dry dichloromethane.
- Prepare a 1.0 M solution of potassium tert-butoxide in tetrahydrofuran.
- Add the FAME extract and 0.2 ml 3-pyridinemethanol to 0.1 ml of solution made in step 5.2.
- Heat the solution at 40 °C for 30 min in a closed vial.
- After cooling to room temperature, add purified deionized water (2 ml, see Materials Table) and hexane (4 ml). Mix with a vortex, allow phase to separate, and collect the organic phase.
- Dry it by adding anhydrous sodium sulfate until the organic phase is perfectly clear. Transfer it into a clean tube. Then evaporate to 200 µl. Transfer the sample into a GC vial with insert.
6. DMOX Preparation from FAME17
- Evaporate the FAME extract from Section 3 with a flow of nitrogen (at least 10 mg dry material).
- To the FAME dry extract, add 250 mg of 2-amino-2-methyl-1-propanol. Flush the vessel with nitrogen, add a stopper, and place it in a heating block overnight at 190 °C.
- After cooling to room temperature, add 3 ml dichloromethane to the tube, and 5 ml purified deionized water (See Materials Table).
- Shake for phase separation and then remove the aqueous phase.
- Wash the organic phase with 5 ml water. Shake for phase separation and then remove the aqueous phase.
- Dry by adding anhydrous sodium sulfate until the organic phase is perfectly clear and transfer it into a clean tube. Evaporate under a stream of nitrogen until reaching a volume of 200 µl. Transfer the sample into a GC vial with insert.
Results
The strategy of FA identification from bacterial cells is presented in Figure 1. Each step provides complementary spectral information or information about chromatographic retention. Step 1 consists of preliminary FA identification using a standard solution. Step 2 allows for the interpretation of FAME EI spectra and their ECL, in order to tentatively identify the products. Step 3 identifies the exact branching location in branched chain-FAs. Finally, step 4 identifies th...
Discussion
The FAs chromatogram profiles shown in Table 1 correspond to B. cereus ATCC 14579 grown on an agar plate surface. Similar profiles were obtained when the bacterium was grown in aerated liquid media at the same temperature8. In the case of bacteria grown in liquid media, the bacterial biomass is collected by centrifugation of the growth medium and can be washed according to previously described protocols depending on the growth conditions8,19. The identification of the vario...
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgements
Authors are grateful to Thomas Mison for his technical support, and to Rachel Kopec for revising the manuscript.
Materials
| Name | Company | Catalog Number | Comments |
| GC/MS | Shimadzu | QP2010 | |
| capillary column ZB WAX | Phenomenex | 7HG-G007-11 | 30 m x 0.25 mm x 0.25 µm |
| Methanol Lichrosolv | VWR | 1.06018.2500 | |
| potassium hydroxide | Aldrich | P1767 | |
| THF | Hipersolv Chromanorm | 28559.320 | |
| Dichloromethane | Hipersolv Chromanorm | 23373.320 | |
| Hexane | Hipersolv Chromanorm | 24575.320 | |
| 3-pyridinemethanol | Aldrich | P6-680-7 | |
| potassium tertiobutoxide | Aldrich | 156671 | |
| 2-amino-2-methyl-1-propanol | A-9879 | ||
| MilliQ Academic | Millipore | ZMQS50001 | |
| Bacterial Acid Methyl Ester (BAME) Mix | Sigma-Aldrich | 47080-U Supelco |
References
- Christie, W. W., Han, X. . Lipid Analysis 4th Edition. , (2010).
- HÜbschmann, H. -. J. . Handbook of GC-MS: fundamental and application. Third edition. , (2015).
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. MIDI Technical note. 101, 1-6 (1990).
- Spitzer, V. Structure analysis of fatty acids by gas chromatography - Low resolution electron impact mass spectrometry of their 4,4-dimethyloxazoline derivatives - A review. Prog Lipid Res. 35 (4), 387-408 (1996).
- Harvey, D. J., Christie, W. W. . Advances in lipid methodology. Volume 1. , 19-80 (1992).
- Diomande, S. E., Nguyen-The, C., Guinebretière, M. -. H., Broussolle, V., Brillard, J. Role of fatty acids in Bacillus environmental adaptation. Front Microbiol. 6, (2015).
- Brillard, J., et al. Identification of Bacillus cereus Genes Specifically Expressed during Growth at Low Temperatures. Appl Environ Microbiol. 76 (8), 2562-2573 (2010).
- de Sarrau, B., et al. Influence of Anaerobiosis and Low Temperature on Bacillus cereus Growth, Metabolism, and Membrane Properties. Appl Environ Microbiol. 78 (6), 1715-1723 (2012).
- Diomandé, S. E., et al. Involvement of the CasK/R two-component system in optimal unsaturation of the Bacillus cereus fatty acids during low-temperature growth. Int J Food Microbiol. 213, 110-117 (2015).
- Berkeley, R. C. W., Heyndrickx, M., Logan, N., De Vos, P., Berkeley, R. C. W. . Applications and Systematics of Bacillus and Relatives. , 1-7 (2002).
- Kämpfer, P. Limits and Possibilities of Total Fatty Acid Analysis for Classification and Identification of Bacillus Species. System. Appl. Microbiol. 17 (1), 86-98 (1994).
- Kaneda, T. Fatty-acids of genus bacillus - example of branched-chain preference. Bacteriol Rev. 41 (2), 391-418 (1977).
- Chazarreta Cifre, L., Alemany, M., de Mendoza, D., Altabe, S. Exploring the Biosynthesis of Unsaturated Fatty Acids in Bacillus cereus ATCC 14579 and Functional Characterization of Novel Acyl-Lipid Desaturases. Appl Environ Microbiol. 79 (20), 6271-6279 (2013).
- Sasser, M., et al. Identification of Bacillus anthracis from culture using gas chromatographic analysis of fatty acid methyl esters. J AOAC Int. 88 (1), 178-181 (2005).
- Schutter, M. E., Dick, R. P. Comparison of fatty acid methyl ester (FAME) methods for characterizing microbial communities. Soil Sci Soc Am J. 64 (5), 1659-1668 (2000).
- Destaillats, F., Angers, P. One-step methodology for the synthesis of FA picolinyl esters from intact lipids. J Am Oil Chem Soc. 79 (3), 253-256 (2002).
- Fay, L., Richli, U. Location of double-bonds in polyunsaturated fatty-acids by gas-chromatography mass-spectrometry after 4,4-dimethyloxazoline derivatization. J Chromatogr. 541 (1-2), 89-98 (1991).
- Zhang, J. Y., Yu, Q. T., Liu, B. N., Huang, Z. H. Chemical modification in mass spectrometry IV-2-alkenyl-4,4-dimethyloxazolines as derivatives for the double bond location of long-chain olefinic acids. Biol Mass Spect. 15 (1), 33-44 (1988).
- de Sarrau, B., et al. Unsaturated fatty acids from food and in the growth medium improve growth of Bacillus cereus under cold and anaerobic conditions. Food Microbiol. 36 (2), 113-122 (2013).
- Miwa, T. K., Mikolajczak, K. L., Earle, F. R., Wolff, I. A. Gas chromatographic characterization of fatty acids.Identification constants for mono- and dicarboxylic methyl esters. Anal Chem. 32 (13), 1739-1742 (1960).
- van Den Dool, H., Kratz, P. A generalization of the retention index system including linear temperature programmed gas-liquid partition chromatography. J Chromatogr A. 11, 463-471 (1963).
- Stransky, K., Jursik, T., Vitek, A. Standard equivalent chain length values of monoenic and polyenic (methylene interrupted) fatty acids. J High Res Chromatogr. 20 (3), 143-158 (1997).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved