A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
PIP-on-a-chip: A Label-free Study of Protein-phosphoinositide Interactions
In This Article
Summary
Here we present a supported lipid bilayer in the context of a microfluidic platform to study protein-phosphoinositide interactions using a label-free method based on pH modulation.
Abstract
Numerous cellular proteins interact with membrane surfaces to affect essential cellular processes. These interactions can be directed towards a specific lipid component within a membrane, as in the case of phosphoinositides (PIPs), to ensure specific subcellular localization and/or activation. PIPs and cellular PIP-binding domains have been studied extensively to better understand their role in cellular physiology. We applied a pH modulation assay on supported lipid bilayers (SLBs) as a tool to study protein-PIP interactions. In these studies, pH sensitive ortho-Sulforhodamine B conjugated phosphatidylethanolamine is used to detect protein-PIP interactions. Upon binding of a protein to a PIP-containing membrane surface, the interfacial potential is modulated (i.e. change in local pH), shifting the protonation state of the probe. A case study of the successful usage of the pH modulation assay is presented by using phospholipase C delta1 Pleckstrin Homology (PLC-δ1 PH) domain and phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) interaction as an example. The apparent dissociation constant (Kd,app) for this interaction was 0.39 ± 0.05 µM, similar to Kd,app values obtained by others. As previously observed, the PLC-δ1 PH domain is PI(4,5)P2 specific, shows weaker binding towards phosphatidylinositol 4-phosphate, and no binding to pure phosphatidylcholine SLBs. The PIP-on-a-chip assay is advantageous over traditional PIP-binding assays, including but not limited to low sample volume and no ligand/receptor labeling requirements, the ability to test high- and low-affinity membrane interactions with both small and large molecules, and improved signal to noise ratio. Accordingly, the usage of the PIP-on-a-chip approach will facilitate the elucidation of mechanisms of a wide range of membrane interactions. Furthermore, this method could potentially be used in identifying therapeutics that modulate protein's capacity to interact with membranes.
Introduction
Myriad interactions and biochemical processes take place on two-dimensionally fluid membrane surfaces. Membrane-enclosed organelles in eukaryotic cells are unique not only in biochemical processes and their associated proteome but also in their lipid composition. One exceptional class of phospholipids is phosphoinositides (PIPs). Even though they comprise only 1% of the cellular lipidome, they play a crucial role in signal transduction, autophagy, and membrane trafficking, among others1,2,3,4. Dynamic phosphorylation of the inositol head group by cellular PIP kinases gives rise to seven PIP headgroups that are mono-, bis-, or tris-phosphorylated5. Additionally, PIPs define the subcellular identity of membranes and serve as specialized membrane docking sites for proteins/enzymes containing one or more phosphoinositide-binding domains, for example, Pleckstrin Homology (PH), Phox Homology (PX), and epsin N-terminal Homology (ENTH)6,7. One of the best-studied PIP-binding domains is phospholipase C (PLC)-δ1 PH domain that specifically interacts with phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) within a high nanomolar-low micromolar range affinity8,9,10,11.
A variety of qualitative and quantitative in vitro methods have been developed and used to study the mechanism, thermodynamics, and specificity of these interactions. Among the most commonly used PIP-binding assays are surface plasmon resonance (SPR), isothermal calorimetry (ITC), nuclear magnetic resonance (NMR) spectroscopy, liposome flotation/sedimentation assay, and lipid-blots (Fat-blots/PIP-strips)12,13. Even though these are extensively utilized, they all have many disadvantages. For example, SPR, ITC, and NMR require large amounts of sample, expensive instrumentation, and/or trained personnel12,13. Some assay formats such as antibody-based lipid-blots utilize water soluble forms of PIPs and present them in a nonphysiological manner12,14,15,16. In addition, lipid-blots cannot be quantitated reliably and they have often resulted in false positive/negative observations12,17,18. To overcome these challenges and improve upon the current tool set, a new label-free method was established based on a supported lipid bilayer (SLB) in the context of a microfluidic platform, which was successfully applied to the study of protein-PIP interactions (Figure 1)19.
The strategy employed for detecting protein-PIP interactions is based on pH modulation sensing. This involves a pH-sensitive dye that has ortho-Sulforhodamine B (oSRB) directly conjugated to phosphatidylethanolamine lipid head group20. The oSRB-POPE probe (Figure 2A) is highly fluorescent at low pH and quenched at high pH with a pKa around 6.7 within 7.5 mol% PI(4,5)P2-containing SLBs (Figure 5B). PLC-δ1 PH domain has been used extensively for validating protein-PIP-binding methodologies due to its high specificity towards PI(4,5)P2 (Figure 5A)21,22,23,24,25.Hence, we reasoned that the PLC-δ1 PH domain can be used to test its binding to PI(4,5)P2 through the PIP-on-a-chip assay. The PH domain construct used in this study has a net positive charge (pI 8.4), and thus attracts OH- ions (Figure 5C). Upon binding to PI(4,5)P2-containing SLBs, the PH domain brings the OH- ions to the membrane surface, which in turn modulates the interfacial potential and shifts the protonation state of oSRB-POPE (Figure 5C)26. As a function of the PH domain concentration, the fluorescence is quenched (Figure 6A). Finally, the normalized data is fit to a binding isotherm to determine the affinity of the PH domain-PI(4,5)P2 interaction (Figure 6B, 6C).
In this study, a detailed protocol is provided to perform protein binding to PIP-containing SLBs within a microfluidic platform. This protocol takes the reader from assembling the microfluidic device and vesicle preparation to SLB formation and protein binding. In addition, directions for data analysis to extract affinity information for the PLC-δ1 PH domain-PI(4,5)P2 interaction are provided.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Cleaning the Glass Coverslips
- Dilute 7x Cleaning Solution (see Materials Table) 7-fold with deionized water in a 100 mm deep borosilicate glass dish with a flat bottom and heat it up to 95 °C on a level hot plate for 20 min or until the cloudy solution becomes clear.
NOTE: The solution will be hot, use caution to avoid bodily injuries. The 7x Cleaning Solution is a proprietary blend of hexasodium [oxido-[oxido(phosphonatooxy)phosphoryl]oxyphosphoryl] phosphate, 2-(2-butoxyethoxy)ethanol, sodium 1,4-bis(2-ethylhexoxy)-1,4-dioxobutane-2-sulfonate and non-hazardous additions. - Using tweezers, arrange the coverslips (40 mm length x 22 mm width x 0.16-0.19 mm height) on an aluminum staining rack in alternating directions. Keep an empty spot between each coverslip to prevent them from touching each other.
- Fully submerge the staining rack with the coverslips in the cleaning solution. Keep the coverslips in the solution for 1 h.
NOTE: As the water evaporates, keep adding fresh deionized water to keep the solution concentration unchanged. - Take the staining racks out of the cleaning solution and wash the coverslips with copious amounts of deionized water to remove the detergent.
- Dry the coverslips with nitrogen gas (approximately 5 min per staining rack), and then place the coverslips in a kiln for 6 h at 550 °C for annealing.
NOTE: This is a crucial step that smoothens rough features on the glass surface. - Place the coverslips in a plastic container and keep them away from dust.
2. Fabricating Micropatterned PDMS Blocks
- Mix polydimethylsiloxane (PDMS) prepolymer and the curing agent at 10:1 (w/w) ratio in a large plastic weigh boat and degas it in vacuum for 1 h with vacuum strength at 500 Torr or less.
NOTE: PDMS is a transparent and inert silicone polymer. To get the desired PDMS block thickness (0.5 cm), mix 55 g of the prepolymer with 5.5 g of the curing agent. - Place the silicon master which contains multiple replicates of the same SU-8 micropattern in a large (10 cm base diameter) plastic weigh boat and pour in the degassed PDMS. Then, cure it in a dry oven at 60 °C overnight.
NOTE: Micropatterns are large enough to be seen with the naked eye. Each pattern contains 8 microchannels (100 µm width x 40 µm height x 1 cm length) with 40 µm spacing in between (Figure 1A, 1B). Silicon wafer mold comprised of hard baked SU-8 photoresist was fabricated in a nanofabrication facility27. Other approaches for master preparation include hydrofluoric acid (HF) etching, high-temperature water etching, xurography, and 3D printing28,29,30,31. Commercial sources for silicon master preparation can also be used. The patterns for the microfluidic device was designed with a drafting software (see table of materials). The original file containing the pattern design is provided as a supplemental file "Pattern Design.dwg", which can be provided directly to the manufacturer. - Gently, peel off the PDMS from the silicon master with hands. Mark the boundaries of each micropattern in rectangles using a surgical scalpel and a ruler. Then, cut the PDMS into blocks.
- Punch 16 holes (8 inlets and 8 outlets per block) at both ends of each microchannel with a biopsy punch (1.0 mm hole diameter) as indicated in Figure 3B.
- Apply tape over each PDMS block to protect the micropatterns from damage and dust. Store them in a plastic container.
3. Preparing Small Unilamellar Vesicles (SUVs)
NOTE: Negative control bilayer composition is 99.5 mol% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 0.5 mol% ortho-Sulforhodamine B-1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (oSRB-POPE). Test bilayer composition is 92.0 mol% POPC, 0.5 mol% oSRB-POPE, and 7.5 mol% of either L-α-phosphatidylinositol-4-phosphate (PI4P) or L-α-phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2). Below is the procedure for preparing 92.0 mol% POPC, 0.5 mol% oSRB-POPE, and 7.5 mol% PI(4,5)P2-containing SUVs. The synthesis of oSRB-POPE used in this study was previously described20.
- Calculate the volume of POPC, PI(4,5)P2, and oSRB-POPE that needs to mixed, which accounts for the following total amounts: 2.22 mg of POPC, 0.26 mg of PI(4,5)P2, and 2.1 x 10-5 mg of oSRB-POPE.
- Pipette the calculated volume of POPC, PI(4,5)P2, oSRB-POPE into a single 20 mL glass scintillation vial.
NOTE: POPC and oSRB-POPE stock solutions come in a chloroform solution, whereas the PI(4,5)P2 (and other phosphoinositides) stock comes in a chloroform:methanol:water (20:9:1) solvent mixture. For accuracy, wet the pipette tip with chloroform before using it. For safety, this step should take place inside a chemical fume hood. - Dry the mixture in a stream of nitrogen gas inside a chemical fume hood for 10 min or until the solvent evaporates and a thin lipid film forms at the bottom of the vial.
- Desiccate the mixture under vacuum for ≥3 h at a vacuum strength of 10 mTorr to remove any residual organic solvent.
NOTE: The 3 h desiccation time is sufficient for the scale of SUV preparation described in this protocol, which is adequate for 50 experiments. If a larger scale of SUV preparation is required, overnight desiccation can be performed. - Rehydrate the dried lipid film with 5 mL of running buffer (20 mM HEPES, 100 mM NaCl, at pH 7.0) and place it in an ultrasonic bath at an operating frequency of 35 kHz for 30 min at RT.
NOTE: Mixing the amounts of lipids specified in step 3.1 and adding 5 mL of running buffer in step 3.5 yields a vesicle suspension at 0.5 mg/mL. At this stage, the vesicle suspension is a mix of unilamellar and multilamellar vesicles (Figure 2B). The solution will appear pink/purple in color and relatively turbid. - Freeze-thaw the vesicle suspension with liquid nitrogen and water bath at 40 °C to get unilamellar vesicles. Repeat the freeze-thaw 10 times.
NOTE: (Optional) To ensure the absence of non-unilamellar vesicles in the suspension, centrifugation step can be applied32,33. - Extrude the vesicle suspension through a 0.10 µm track-etched polycarbonate membrane using a lipid extruder (see table of materials) to enrich for SUVs. Repeat the extrusion 10 times.
NOTE: A 15-mL conical tube can be used to collect the extruded vesicles. The solution should be void of turbidity at this stage. SUV diameter can be confirmed via dynamic light scattering (DLS) (Figure 2C). SUVs are best suited for forming SLBs34. - Cover the conical tube with aluminum foil and store at 4 °C until ready to use.
4. Assembling the Microfluidic Device
- Test the inlets and outlets of the PDMS block for blockage by squirting deionized water through the holes using a water wash bottle. Then, dry the PDMS block with nitrogen gas.
NOTE: This step will also remove any dust particles that might have been trapped within and/or between channels. If necessary, dedust pre-cleaned coverslips with nitrogen gas to remove any dust particles. - Place the PDMS block and the pre-cleaned coverslip (see section 1) inside the oxygen plasma system (see table of materials) sample chamber to expose with oxygen plasma for 45 s with power setting at 75 Watts, oxygen flow speed at 10 cm3/min, and vacuum strength at 200 mTorr.
- Place the patterned surface of the PDMS block in contact with the coverslip immediately after the oxygen plasma treatment. Press gently to remove any air bubbles at contact sites.
- Place the device on a level hot plate at 100 °C for 3 min to enhance the bonding.
- Use a wet lint-free wipe (e.g. kimwipe) with 100% ethanol to remove any dust particles from the top (PDMS) and the bottom (glass) of the device. Then, tape the device on top of a glass microscope slide.
NOTE: Do not use an excessive amount of ethanol and avoid ethanol getting into the inlet and outlet channels. Performing this step while the device is still hot is recommended to ensure that the ethanol evaporates immediately. Glass microscopy slides can be stored in 100% ethanol solution to remove dust and other contaminants.
5. Forming Supported Lipid Bilayers (SLBs)
- Transfer 100 µL of PI(4,5)P2-containing SUVs from step 3.8 into a 0.65 mL microcentrifuge tube. Adjust the pH of the solution to ~3.2 by adding 6.4 µL of 0.2 N hydrochloric acid.
NOTE: Confirm the pH of the solution with a pH meter equipped with a micro pH probe (see table of materials). - Pipette 10 µL of the pH-adjusted SUV solution into each channel through the inlet and apply pressure through the pipette until the solution reaches the outlet. Detach the tip from the pipette and leave it attached to the device.
- Repeat the above step for each channel and then incubate the device for 10 min at RT.
NOTE: Injection of vesicles into microchannels should be performed immediately after the device assembly. - Cut sets of inlet and outlet tubing each 60 cm (inlet tubing) and 8 cm (outlet tubing) long, respectively.
NOTE: Cutting the tubing diagonally and creating a sharp edge, makes it easier to insert the tubing into the inlets and outlets. The outlet tubing should have an arc shape. The length of the inlet tubing may vary based on the microscope setup. The internal diameter of the tubing is 0.05 cm. - Using tweezers, connect the outlet tubing set to the device, and then tape the device onto a microscope stage.
- Submerge one end of the inlet tubing set in 25 mL of running buffer contained in a conical tube and tape it to make sure that the tubing is secured.
- Place the conical tube on a higher ground (~20 cm) than the device in order to push the solution through the microchannels via gravity flow; a lab jack can be used to achieve this.
- For each inlet tube, use a syringe to draw 1 mL of running buffer from the free end of the tubing. Remove the pipette tip from the inlet and insert the free end of the inlet tubing into the device.
NOTE: Introduce a drop of running buffer onto the inlet to reduce the probability of an air bubble being introduced into the channel during this step. After the inlet tubing is attached to the device, use a lint-free wipe to remove the excess buffer. - Repeat the above step to connect all the inlet tubing pieces to the device.
NOTE: Flowing running buffer through the channels helps to remove excess vesicles and equilibrate the bilayer to experimental conditions. - Open the microscope control software (see table of materials). On the left panel, click on the "Microscope" tab and choose the "10X" objective.
- Click on "Live" and then "Alexa 568" image icons on the toolbar. Using the fine and course adjustment knobs, focus on the microchannels.
- Scan through the device to check the quality of the SLBs and the channels (Figure 8). Then, click "FL Shutter Closed" image icon on the toolbar.
- Click on the "Acquisition" tab, and under "Basic adjustments" select "Exposure time." Set the exposure time to "200 ms".
- On the left panel, click on "Multidimensional Acquisition" and under the filters menu select the red channel (marked as "Alexa 568"). Then, click on the "time lapse" menu, set the time interval to 5 min, duration to 30 min, and click "Start".
NOTE: Based on the duration (30 min) and the flow rate (~1.0 µL/min), 30 µL of running buffer is used to equilibrate the SLB within a channel, i.e. 240 µL of running buffer is used for all 8 channels. - Select the "Circle" tool under the "Measure" tab and draw a circle in any channel. Right click while the circle is selected and choose "Properties". Under the "Profile" tab, check "all T" to view the fluorescence intensity as a function of time.
- Make sure this curve reaches a plateau, which indicates equilibrium, before proceeding to the next step.
- Lower the buffer solution to an equal ground as the device to stop the flow.
- Save the time lapse file.
6. Testing PLC-δ1 PH domain interaction with PI(4,5)P2-containing SLBs
- Prepare dilutions of the PH domain using the running buffer as a diluent.
NOTE: Since there are eight channels within the device, use at least two of them for blank controls. Pick the far ends on each side and use the remaining channels for protein dilutions. The following PH domain concentrations were tested: 0.10, 0.25, 0.50, 1.00, and 2.50 µM. Approximately 200 µL of each dilution was sufficient to reach equilibrium within 30 min. - One at a time, detach each outlet tubing and apply 200 µL of each protein dilution into the outlet channel using a pipette. Do not apply any pressure, let gravity do the work. Detach the tip from the pipette and leave it attached to the microfluidic device.
- Repeat the above step for each channel and make sure that air bubbles are not introduced into the channels during this process.
- Lower the inlet tubing to a ground below the microfluidic device to start flowing the protein through the microchannels. Tape the free end of the tubing to a waste container. Flow the protein dilutions for 30 min.
NOTE: This will reverse the flow and allow the protein to be introduced into the channels. The time to equilibration and volume of protein dilutions needed will be dependent on the affinity of the interaction. - On the left panel of the software, under the "Time Lapse" tab, click on "Start" to begin imaging again.
- Repeat the step 5.15 to make sure equilibrium is reached. When the experiment is complete, save the time lapse file.
7. Assessing Membrane Fluidity
NOTE: Fluorescence Recover After Photobleaching (FRAP) experiments should be performed with each new batch of SUVs and cleaned glass coverslips to ensure that the SLBs are fluid.
- Follow the procedure for cleaning glass coverslips (1.1-1.6), fabricating micropatterned PDMS blocks (2.1-2.5), preparing SUVs (3.1-3.8), assembling the microfluidic device (4.1-4.5) and forming SLBs (5.1-5.17) as previously described.
- Focus the microscope on the bilayer, set the exposure time to 200 ms, and the binning to 1.
- Photo-bleach a circular spot using a 532 nm, 2 mW laser beam (13 µM radius). Immediately after photobleaching, start capturing a series of images every 3 s for the first 45 s, followed by a 30 s interval for the remainder of the time (15 min total duration). Once the data acquisition is complete, save the time lapse file.
- Select the bleached area using the circle drawing tool to extract fluorescence intensity values as a function of time. Additionally, select two more areas nearby, one corresponding to an unbleached region and one corresponding to a region without any SLBs.
- Calculate the fluorescence recovery (y) using the following equation:
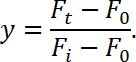
NOTE: Here Ft represents the intensity of the bleached region as a function of time, Fi represents the fluorescence intensity before bleaching (use "1" as a normalized value in this case); F0 is the background intensity. - Plot fluorescence recovery (y-axis) as a function of time (x-axis) to generate a FRAP curve. Then, fit the data to a single exponential function as follows,
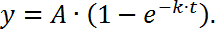
NOTE: Here A represents the mobile fraction and k is the kinetic constant for the mobile fraction, which is used to calculate the half-time to recovery (t1/2):

- Use the t1/2 to calculate the diffusion constant (D):

NOTE: Here R represents the laser beam radius (13 µm). The diffusion constant for a fluid bilayer should be ≥1.0 µm2/s.
8. Processing Data
NOTE: The routine of the data analysis will be dependent on the microscope, image processing software, and the curve-fitting software being used.
- Open the time lapse files of before (from step 5.17) and after (from step 6.6) titrating the PH domain. Go to the last frame of each time lapse file and do a line scan across all the microchannels to obtain the fluorescence intensity data as a function of distance in pixels (Figure 6A). Transfer the data to a spreadsheet software.
- For each microchannel (before and after PH domain addition), sample some data within the channel and either side of the channel representing the baseline (Figure 7A).
NOTE: The blank channel fluorescence intensity should have remained unchanged. All the other channels are normalized to the blank channel, so it is crucial to have two blank channels and make sure that their fluorescence intensities remain consistent with each other. - Apply the formula below to normalize the data to the blank channel; a sample calculation is provided in Figure 7B.

NOTE: Here ΔFprotein represents the background subtracted fluorescence intensity of the protein channel, ΔFblank represents the background subtracted fluorescence intensity of the blank channel, pre and post refer to before and after protein titration steps, and α represents the correction factor which is the ratio of the fluorescence intensities of the protein channel and the blank channel ([Fprotein/Fblank]pre) at a pre-protein titration step. Since the fluorescence intensity decreases as a function of the PH domain concentration, normalized data will be negative in value. - Using a curve-fitting software (see Materials Table), plot the normalized fluorescence as a function of PH domain concentration and fit to a Langmuir isotherm (Figure 6C):
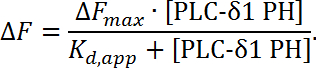
NOTE: Here ∆F represents the change in fluorescence intensity relative to the maximum change in fluorescence intensity when a saturation concentration of protein is present (∆Fmax), whereas the Kd,app represents the apparent dissociation constant, which is equal to the bulk protein concentration ([PLC-δ1 PH]) at which 50% coverage (membrane bound complex) is achieved. This is an equilibrium-based binding measurement so the normalized data is fit to a simple Langmuir isotherm to extract an apparent experimental parameter Kd,app. Based on the fitting software the Kd,app for PLC-δ1 PH-PI(4,5)P2 interaction is 0.39 ± 0.05 µM (Figure 6B).
Access restricted. Please log in or start a trial to view this content.
Results
We used the pH modulation assay to study the PLC-δ1 PH domain-PI(4,5)P2 interaction within a PIP-on-a-chip microdevice (Figure 1). Through a detailed protocol, we demonstrated how to prepare and assemble microfluidic device components, make small unilamellar vesicles (SUVs) (Figure 2), form SLBs within a device (Figure 3), and tested protein binding to PIP-containing SLBs. A flowchart...
Access restricted. Please log in or start a trial to view this content.
Discussion
Each PIP variant, albeit at low concentrations, is present on the cytosolic surface of specific organelles where they contribute to the establishment of a unique physical composition and functional specificity of the organellar membrane1. One of the most important uses of PIPs is as a specific docking platform for the multitude of proteins requiring specific subcellular localization and/or activation6,7. Due to their role in cellular physi...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
D.S. and C.E.C. were supported, in part, by grant AI053531 (NIAID, NIH); S.S and P.S.C. were supported by grant N00014-14-1-0792 (ONR).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Coverslip | |||
| Glass Coverslips: Rectangles | Fisher Scientific | 12-544B | 22 x 40 x 0.16 - 0.19 mm, No. 1 1/2; Borosilicate Glass |
| 7X Cleaning Solution | MP Biomedicals | 976670 | Detergent |
| PYREX Crystallizing Dish | Corning | 3140-190 | Borosilicate glass dish with a flat bottom; Diameter x Height (190 x 100 mm); Distributor: VWR (89090-700) |
| Sentry Xpress 2.0 | Paragon Industries | SC-2 | Kiln |
| Name | Company | Catalog Number | Comments |
| PDMS | |||
| Sylgard 184 Silicone Elastomer Kit | Dow Corning | 4019862 | Polydimethylsiloxane (PDMS); Distributor: Ellsworth Adhesives |
| PYREX Desiccator | VWR | 89134-402 | Vacuum Rated |
| Biopsy punch | Harris | 15110-10 | Harris Uni-Core; 1.0 mm diameter; Miltex Biopsy Punch with Plunger (Cat. No. 15110-10) can be used as an alternative |
| Name | Company | Catalog Number | Comments |
| Device | |||
| Plasma Cleaning System | PlasmaEtch | PE25-JW | 2-stage Direct Drive Oil Vacuum Pump, O2 service (Krytox Charged) |
| Digital Hot Plate | Benchmark | H3760-H | Purchased through Denville Scientific (Cat. No. 1005640) |
| Frosted Micro Slides | VWR | 48312-003 | Frosted, Selected, and Precleaned; Made of Swiss Glass; Thickness: 1 mm; Dimensions: 75 x 25 mm; GR 144 |
| Name | Company | Catalog Number | Comments |
| Mold | |||
| AutoCAD | Autodesk | v.2016 | Drafting software for the photomask design |
| Photomask | CAD/Art Services | N/A | Design with black background and clear features was printed at 20k dpi resolution on a transparent mask (5 x 7 in) by CAD/Art Services |
| Silicone Wafers | University Wafer | 1575 | Prime Grade, Single Side Polished; 100 mm (4 inch) Diameter; 525 um Thickness |
| SU-8 50 | MicroChem Corp. | N/A | Negative Tone Photoresist; Penn State Nanofabrication Facility Property |
| SU-8 Developer | MicroChem Corp. | N/A | Penn State Nanofabrication Facility Property |
| Name | Company | Catalog Number | Comments |
| SUV | |||
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine | Avanti Polar Lipids | 850457C | POPC |
| L-α-phosphatidylinositol-4-phosphate | Avanti Polar Lipids | 840045X | PI4P |
| L-α-phosphatidylinositol-4,5-bisphosphate | Avanti Polar Lipids | 840046X | PI(4,5)P2 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | 850757C | POPE; Required for the synthesis of oSRB-POPE |
| Lissamine Rhodamine B Sulfonyl Chloride (mixed isomers) | ThermoFisher Scientific | L-20 | Required for the synthesis of oSRB-POPE |
| pH Sensitive Fluorescent Lipid Probe (oSRB-POPE) | In-house | N/A | In-house Synthesis (Huang D. et al. 2013) |
| Glass Scintillation Vial | VWR | 66022-065 | 20 mL volume capacity |
| Aquasonic 250D | VWR | N/A | Ultrasonic Water Bath |
| Nuclepore Track-Etched Membranes | Whatman | 110605 | Polycarbonate Membrane; Diameter: 25 mm; Pore Size: 0.1 um; Distributor: Sigma-Aldrich |
| Chloroform | VWR | CX1054-6 | HPLC grade |
| LIPEX Extruder | Transferra Nanosciences | T.001 | LIPEX 10 mL Thermobarrel Extruder |
| Viscotek 802 DLS | Malvern Instruments | N/A | Dynamic Light Scattering; Penn State X-Ray Crystallography Facility Property |
| Name | Company | Catalog Number | Comments |
| Data Analysis | |||
| GraphPad Prism | GraphPad Software | v.6 | Curve-fitting software for data analysis |
| Name | Company | Catalog Number | Comments |
| Microscope | |||
| Axiovert 200M Epifluorescence Microscope | Carl Zeiss Microscopy | N/A | Microscope |
| AxioCam MRm Camera | Carl Zeiss Microscopy | N/A | Camera |
| X-Cite 120 | Excelitas Technologies | N/A | Light Source |
| Alexa 568 Filter Set | Carl Zeiss Microscopy | N/A | Ex/Em 576/603 nm |
| AxioVision LE64 v.4.9.1.0 Software | Carl Zeiss Microscopy | N/A | Image Processing Software |
| Name | Company | Catalog Number | Comments |
| Other | |||
| Tips | VWR | 10034-132 | 200 uL pipette tips; Thin and smooth tip for applying the protein solution into the microfluidic channel |
| Tips | VWR | 53509-070 | 10 uL pipette tips; Thin and smooth tip for applying the vesicle solution into the microfluidic channel |
| Orion Star A321 pH meter | Thermo Scientific | STARA3210 | pH meter |
| Orion micro pH probe | Thermo Scientific | 8220BNWP | micro pH probe |
| N-(2-Hydroxyethyl)-Piperazine-N'-(2-Ethanesulfonic Acid) | VWR | VWRB30487 | HEPES, Free Acid |
| Sodium Chloride | VWR | BDH8014-2.5KGR | NaCl |
| Tubing | Allied Wire & Cable | TFT-200-24 N | Internal Diameter: 0.020-0.026 inches (0.051-0.066 cm); Wall Thickness: 0.010 inches (0.025 cm); Flexible Polytetrafluoroethylene Thin-Wall Tubing; Natural Color |
| Nitrogen Gas - Industrial | Praxair | N/A | Local Provider |
| Oxygen Gas - Industrial | Praxair | N/A | Local Provider |
| Liquid Nitrogen | Praxair | N/A | Local Provider |
References
- Di Paolo, G., De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443 (7112), 651-657 (2006).
- Shewan, A., Eastburn, D. J., Mostov, K. Phosphoinositides in cell architecture. Cold Spring Harb Perspect Biol. 3 (8), a004796(2011).
- Picas, L., Gaits-Iacovoni, F., Goud, B. The emerging role of phosphoinositide clustering in intracellular trafficking and signal transduction. F1000Res. 5, (2016).
- Lystad, A. H., Simonsen, A. Phosphoinositide-binding proteins in autophagy. FEBS Lett. 590 (15), 2454-2468 (2016).
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol Rev. 93, 1019-1137 (2013).
- Lemmon, M. A. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 9 (2), 99-111 (2008).
- Kutateladze, T. G. Translation of the phosphoinositide code by PI effectors. Nat Chem Biol. 6 (7), 507-513 (2010).
- Harlan, J. E., Hajduk, P. J., Yoon, H. S., Fesik, S. W. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 371 (6493), 168-170 (1994).
- Garcia, P., et al. The pleckstrin homology domain of phospholipase C-delta 1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry. 34 (49), 16228-16234 (1995).
- Lemmon, M. A., Ferguson, K. M., O'Brien, R., Sigler, P. B., Schlessinger, J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci U S A. 92 (23), 10472-10476 (1995).
- Flesch, F. M., Yu, J. W., Lemmon, M. A., Burger, K. N. Membrane activity of the phospholipase C-delta1 pleckstrin homology (PH) domain. Biochem J. 389, 435-441 (2005).
- Narayan, K., Lemmon, M. A. Determining selectivity of phosphoinositide-binding domains. Methods. 39 (2), 122-133 (2006).
- Scott, J. L., Musselman, C. A., Adu-Gyamfi, E., Kutateladze, T. G., Stahelin, R. V. Emerging methodologies to investigate lipid-protein interactions. Integr Biol (Camb). 4 (3), 247-258 (2012).
- Dowler, S., Currie, R. A., Downes, C. P., Alessi, D. R. DAPP1: A dual adaptor for phosphotyrosine and 3-phosphoinositides. Biochem J. 342, 7-12 (1999).
- He, J., et al. Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J Biol Chem. 286 (21), 18650-18657 (2011).
- Ceccarelli, D. F., et al. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J Biol Chem. 282 (18), 13864-13874 (2007).
- Huang, S., Gao, L., Blanchoin, L., Staiger, C. J. Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol Biol Cell. 17 (4), 1946-1958 (2006).
- Yu, J. W., et al. Genome-eide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 13 (5), 677-688 (2004).
- Jung, H., Robison, A. D., Cremer, P. S. Detecting protein-ligand binding on supported bilayers by local pH modulation. J Am Chem Soc. 131 (3), 1006-1014 (2009).
- Huang, D., Zhao, T., Xu, W., Yang, T., Cremer, P. S. Sensing small molecule interactions with lipid membranes by local pH modulation. Anal Chem. 85 (21), 10240-10248 (2013).
- Saxena, A., et al. Phosphoinositide binding by the pleckstrin homology domains of Ipl and Tih1. J Biol Chem. 277 (51), 49935-49944 (2002).
- Knödler, A., Mayinger, P. Analysis of phosphoinositide-binding proteins using liposomes as an affinity matrix. Biotechniques. 38 (6), 858-862 (2005).
- Baumann, M. K., Swann, M. J., Textor, M., Reimhult, E. Pleckstrin homology-phospholipase C-delta1 interaction with phosphatidylinositol 4,5-bisphosphate containing supported lipid bilayers monitored in situ with dual polarization interferometry. Anal Chem. 83 (16), 6267-6274 (2011).
- Saliba, A. E., et al. A quantitative liposome microarray to systematically characterize protein-lipid interactions. Nat Methods. 11 (1), 47-50 (2014).
- Arauz, E., Aggarwal, V., Jain, A., Ha, T., Chen, J. Single-molecule analysis of lipid-protein interactions in crude cell lysates. Anal Chem. 88 (8), 4269-4276 (2016).
- Best, Q. A., Xu, R., McCarroll, M. E., Wang, L., Dyer, D. J. Design and investigation of a series of rhodamine-based fluorescent probes for optical measurements of pH. Org Lett. 12 (14), 3219-3221 (2010).
- Lee, J., Choi, K. H., Yoo, K. Innovative SU-8 lithography techniques and their applications. Micromachines. 6 (1), 1-18 (2014).
- Poyton, M. F., Sendecki, A. M., Cong, X., Cremer, P. S. Cu(2+) binds to phosphatidylethanolamine and increases oxidation in lipid membranes. J Am Chem Soc. 138 (5), 1584-1590 (2016).
- Karasek, P., Grym, J., Roth, M., Planeta, J., Foret, F. Etching of glass microchips with supercritical water. Lab Chip. 15 (1), 311-318 (2015).
- Thomas, M. S., et al. Print-and-peel fabrication for microfluidics: what's in it for biomedical applications? Ann Biomed Eng. 38 (1), 21-32 (2010).
- Waheed, S., et al. 3D printed microfluidic devices: enablers and barriers. Lab Chip. 16 (11), 1993-2013 (2016).
- Axmann, M., Schutz, G. J., Huppa, J. B. Single molecule fluorescence microscopy on planar supported bilayers. J Vis Exp. (105), e53158(2015).
- Barenholz, Y., et al. A simple method for the preparation of homogeneous phospholipid vesicles. Biochemistry. 16 (12), 2806-2810 (1977).
- Castellana, E. T., Cremer, P. S. Solid supported lipid bilayers: From biophysical studies to sensor design. Surface Science Reports. 61 (10), 429-444 (2006).
- Hamai, C., Yang, T., Kataoka, S., Cremer, P. S., Musser, S. M. Effect of average phospholipid curvature on supported bilayer formation on glass by vesicle fusion. Biophys J. 90 (4), 1241-1248 (2006).
- Tero, R. Substrate effects on the formation process, structure and physicochemical properties of supported lipid bilayers. Materials. 5 (12), 2658-2680 (2012).
- Ferguson, K. M., Lemmon, M. A., Schlessinger, J., Sigler, P. B. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 83 (6), 1037-1046 (1995).
- Simonsson, L., Hook, F. Formation and diffusivity characterization of supported lipid bilayers with complex lipid compositions. Langmuir. 28 (28), 10528-10533 (2012).
- Cong, X., Poyton, M. F., Baxter, A. J., Pullanchery, S., Cremer, P. S. Unquenchable surface potential dramatically enhances Cu(2+) binding to phosphatidylserine lipids. J Am Chem Soc. 137 (24), 7785-7792 (2015).
- Robison, A. D., et al. Polyarginine interacts more strongly and cooperatively than polylysine with phospholipid bilayers. J Phys Chem B. 120 (35), 9287-9296 (2016).
- Robison, A. D., Huang, D., Jung, H., Cremer, P. S. Fluorescence modulation sensing of positively and negatively charged proteins on lipid bilayers. Biointerphases. 8 (1), 1(2013).
- Tabaei, S. R., et al. Formation of cholesterol-rich supported membranes using solvent-assisted lipid self-assembly. Langmuir. 30 (44), 13345-13352 (2014).
- Johnson, S. J., et al. Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons. Biophys J. 59 (2), 289-294 (1991).
- Koenig, B. W., et al. Neutron reflectivity and atomic force microscopy studies of a lipid bilayer in water adsorbed to the surface of a silicon single crystal. Langmuir. 12 (5), 1343-1350 (1996).
- Tanaka, M., Sackmann, E. Polymer-supported membranes as models of the cell surface. Nature. 437 (7059), 656-663 (2005).
- Renner, L., et al. Supported lipid bilayers on spacious and pH-responsive polymer cushions with varied hydrophilicity. J Phys Chem B. 112 (20), 6373-6378 (2008).
- Wagner, M. L., Tamm, L. K. Tethered polymer-supported planar lipid bilayers for reconstitution of integral membrane proteins: Silane-polyethyleneglycol-lipid as a cushion and covalent linker. Biophys J. 79 (3), 1400-1414 (2000).
- Pace, H., et al. Preserved transmembrane protein mobility in polymer-supported lipid bilayers derived from cell membranes. Anal Chem. 87 (18), 9194-9203 (2015).
- Braunger, J. A., Kramer, C., Morick, D., Steinem, C. Solid supported membranes doped with PIP2: Influence of ionic strength and pH on bilayer formation and membrane organization. Langmuir. 29 (46), 14204-14213 (2013).
- Paridon, P. A., de Kruijff, B., Ouwerkerk, R., Wirtz, K. W. Polyphosphoinositides undergo charge neutralization in the physiological pH range: A 31P-NMR study. Biochim Biophys Acta. 877 (1), 216-219 (1986).
- Liu, C., Huang, D., Yang, T., Cremer, P. S. Monitoring phosphatidic acid formation in intact phosphatidylcholine bilayers upon phospholipase D catalysis. Anal Chem. 86 (3), 1753-1759 (2014).
- Saad, J. S., et al. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A. 103 (30), 11364-11369 (2006).
- Hsu, N. Y., et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 141 (5), 799-811 (2010).
- Del Campo, C. M., et al. Structural basis for PI(4)P-specific membrane recruitment of the Legionella pneumophila effector DrrA/SidM. Structure. 22 (3), 397-408 (2014).
- Kolli, S., et al. Structure-function analysis of vaccinia virus H7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication. J Virol. 89 (4), 2209-2219 (2015).
- Cho, N. J., et al. Phosphatidylinositol 4,5-bisphosphate is an HCV NS5A ligand and mediates replication of the viral genome. Gastroenterology. 148 (3), 616-625 (2015).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved