A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Spectral Reflectometric Microscopy on Myelinated Axons In Situ
In This Article
Summary
Here, we present a step-by-step protocol for imaging myelinated axons in a fixed brain slice using a label-free nanoscale imaging technique based on spectral reflectometry.
Abstract
In a mammalian nervous system, myelin provides an electrical insulation by enwrapping the axon fibers in a multilayered spiral. Inspired by its highly-organized subcellular architecture, we recently developed a new imaging modality, named spectral reflectometry (SpeRe), which enables unprecedented label-free nanoscale imaging of the live myelinated axons in situ. The underlying principle is to obtain nanostructural information by analyzing the reflectance spectrum of the multilayered subcellular structure. In this article, we describe a detailed step-by-step protocol for performing a basic SpeRe imaging of the nervous tissues using a commercial confocal microscopic system, equipped with a white-light laser and a tunable filter. We cover the procedures of sample preparation, acquisition of spectral data, and image processing for obtaining nanostructural information.
Introduction
In the mammalian nervous system, myelin provides rapid nerve conduction and axonal integrity by enwrapping the axon fibers with multilayered membranous sheaths. Its multilayered structure is composed of alternating nanoscale thin-films composed of plasma membranes (~5 nm), cytosol (~3 nm), and extracellular spaces (~7 nm)1,2. Optical microscopy, including the recent super-resolution microscopy, are not suitable for observing the nanoscale myelin dynamics due to their insufficient resolution due to optical diffraction3,4,5. Although electron microscopy can provide fine details of the myelin nanostructure, it is not compatible with the living biological systems due to highly invasive sample preparations involving chemical fixation and ultrasectioning6,7. Until recently, there has been no technique applicable to observe nanoscale dynamics of myelinated axons in situ.
Schain et al. previously reported that myelinated axons exhibit colorful light reflectance8. By adopting the spectroscopic analysis on the reflected light, we have devised a new imaging modality for nanoscale imaging of myelinated axons, named spectral reflectometry (SpeRe)9. SpeRe is based on the thin-film interference occurring in the multi-layered structure of the myelin sheath (Figure 1). By optic simulation on various axons, we have revealed that the reflectance spectrum is a periodic function of wavenumber and its periodicity (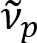 ) is inversely proportional to the axon diameter (d). This simple relationship (
) is inversely proportional to the axon diameter (d). This simple relationship ( ) offers facile quantification of axon diameter from the SpeRe data. Utilizing this, we revealed the prevalent axon bulging under mild traumatic brain injury in our prior report.
) offers facile quantification of axon diameter from the SpeRe data. Utilizing this, we revealed the prevalent axon bulging under mild traumatic brain injury in our prior report.
The SpeRe system is based on confocal microscopy and consists of a specialized laser source and filters (Figure 2). The input source is a white-light laser, providing broadband spectral output visible to infrared regions. For the spectral scan, the system is equipped with two acousto-optic devices: an acousto-optic tunable filter (AOTF) for delivering a selected wavelength from the input broadband source and an acousto-optic beam splitter (AOBS) for guiding the selected reflected wavelength to the detector. The software for hyperspectral confocal microscopy (see Table of Materials) provides a customizable spectral scan option to sequentially acquire the reflectance images at various input wavelengths. In addition, chromatic aberration can critically interfere in the spectral measurement; therefore, use of an apochromat objective lens is recommended.
Of note, white-light lasers produce an uneven spectral output and the optical components also affect the spectral profile. Therefore, the acquired spectra need to be calibrated for the subsequent quantitative analysis. A protected silver mirror is typically used as a reference, which provides a nearly constant reflectance (> 97%) over the full visible region. The acquired spectra are then divided by the reference spectra from the mirror.
The spectral step size for the spectral scan determines the acquisition speed; thus, it needs to be optimized. As a larger axon has a higher spectral period, it requires finer spectral sampling. For example, an axon with a diameter of 10 µm, one of the largest physiologic axons, has a spectral period of ~8 nm. By applying Nyquist sampling criteria, we employed the spectral sampling interval of 4 nm to cover all the physiologic axons in the mouse nervous tissues. This approach typically takes over several seconds for a full spectral scan and thus is not suitable for in vivo applications, where physiologic motion (e.g. respiration and heartbeat) interferes stable spectral acquisition. We previously solved this issue by instrumenting a customized upright microscope, designed to acquire the full spectrum for each point using an array spectrometer (acquisition speed ≈ 30 ms per pixel).
In this report, we describe a detailed protocol on the SpeRe imaging on a fixed brain slice, which can be performed in a commercial hyperspectral microscope (see Table of Materials). Thus, the protocol can be completed by experimenters without expertise in optical instrumentation. We also cover the potential issues and troubleshooting for acquisition and analysis of SpeRe data.
Protocol
All surgical procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Sungkyunkwan University.
1. Sample Preparation
Note: Autoclave all surgical instruments before animal handling. Conduct all surgical performance in a room dedicated to surgical procedures. Sterile surgical gowns and gloves should be worn by all personnel in the surgical room at all times.
- Tissue Fixation
- Prepare two 10 mL syringes each filled with phosphate-buffered saline (PBS) and 4% paraformaldehyde (PFA) in PBS.
- Anesthetize a 7-12-week-old mouse, (C57BL/6J or any mouse lines with either gender) by administering a mixture of zoletil and xylazine (1:1, 20 mg/kg each) or an alternative suitable anesthesia regime (e.g., mixture of 80 mg/kg ketamine and 10 mg/kg xylazine) with an intraperitoneal injection.
- Once the mouse has reached a surgical plane of anesthesia (use the toe pinch-response method), expose the heart by cutting off the skin and the rib cage with scissors.
- Snip the right atrium with small scissors to drain the blood and perfuse the PBS and PFA solution sequentially through the left ventricle with a needle (perfusion rate = 4 mL/min, volume = 10 mL for each solution).
Note: The mouse becomes stiff if the procedure is successfully completed. The use of eye ointment or sterilization is not necessary as the procedure is terminal. The fixation procedure may be skipped. However, this step is recommended to minimize structural deformations including mechanical tissue damage and ischemic axonal swelling. For further details on tissue fixation, refer to the article by Gage, G. J. et al.10
- Preparation of the Brain Slice
- Decapitate the mouse with surgical scissors through the ventral thorax and abdomen.
- Remove the scalp and periosteum using a suitably small scissors until fully exposing the cranium.
- Break the frontal bone and cut the cranium along the sagittal suture from the occipital bone using small scissors with care not to damage the brain.
- Gently remove the cranium and the dura mater sequentially using fine forceps.
- Extract the brain using a soft plastic spoon, taking care not to damage the tissue.
- Rinse and soak the extracted brain in a 4% PFA solution for 30 min at 4 °C.
- Slice the fixed brain into 100 – 150 µm sections using a vibratome.
Note: For further details on the tissue preparation, refer to the article by Segev et al.11. For subsequent procedures, the tissue should be sliced into sections thinner than the thickness of the spacer.
- Mounting of the Brain Slice
- Prepare 1 glass slide and 2 square cover glasses (22 × 22 × 0.17 mm) for each tissue slice.
- Cut one of the square cover glasses into half (two rectangular pieces) by using a glass cutter.
- Attach the two of the halved cover glasses using a super glue on the slide glass.
Note: The space between the two halved glasses should be slightly larger than the size of the tissue slice. - Harvest the brain slice using a brush or a pipette and lay the tissue on the slide glass between the spacer. Take care not to fold the tissue.
- Dispense 100 μL of PBS on the tissue surface.
- Place the other square cover glass on top of the tissue. Avoid inclusion of air bubbles during this stage.
- Seal around the cover glass using a nail polish (or alternatively suitable adhesives) to prevent evaporation of PBS and contamination by dust during the subsequent imaging session.
Note: The experimenter may pause at this stage.
2. Calibration
- Switch on the microscope at least 1 h before the imaging to allow thermal stabilization of the laser source (see Table of Materials). Then, turn on the software for spectral scanning (see Table of Materials) and click the Acquire button on the top of GUI (Figure 3a).
- Turn on the software shutter for the white-light laser (WLL) and photomultiplier tube (PMT) (Figure 3a).
- Select the xyΛ mode on the drop-down list in Acquisition Mode, then, the laser input mode is automatically changed from Constant percent to Constant Power. Uncheck the check-box of Automatic SP Movement. And then, set the spectral window of the input laser to 470–670 nm and the spectral step size to 4 nm on Λ-Excitation Lambda Scan Settings (Figure 3b).
- SET the spectral range of PMT to 450-690 by double-clicking or moving the adjustment bar for spectral range (Figure 3c).
Note: This spectral range should include the complete bandwidth of the input source. - Select a water-immersion objective lens, suitably with a high numerical aperture (NA > 0.7) and give any value to PMT and laser power to activate AOBS configuration and Live Scan button. Switch the optical path by checking reflection in the AOBS Configuration (Figure 3d).
- Mount a reference mirror (see Table of Materials) on the microscope stage. The mirror surface should be faced against the objective lens. If it is not easy to put the mirror on the microscope stage, bond a mirror onto the flat plate (e.g. slide glass).
- Control the microscope stage to align the focal plane to the mirror surface.
- Adjust the PMT gain and the laser power considering the dynamic range of the detector and then change the laser input mode from Constant Percent to Constant Power.
Note: As is typical, a PMT gain of 500 (V) and a relative laser power of 0.1% are used at 570 nm. - Confirm in a pseudo-color mode to check that there is no saturation throughout the wavelength range. If saturation is observed, lower the laser power (Figure 3a).
- Run the Lambda Scan acquisition.
- Remove the mirror from the stage and repeat the same acquisition without a sample in order to obtain the dark reference (i.e., dark offset).
- Save the data in a Multistacked TIF format.
3. SpeRe Image Acquisition
- Place the mounted tissue on the microscope stage. To roughly align the tissue to the focal plane of the objective lens, use the wide-field fluorescence mode through an eye-piece.
- With the Live Scan on, control the microscope stage to align the focal plane to the region of interest in the tissue. To avoid the background noise from the cover slip, select the target region of at least 15 μm depth from the glass-tissue interface.
- Acquire the spectral image stack for the target region using the same procedure as described in steps 2.1–2.10.
- Save the data for the tissue and the dark offset in Multistacked TIF format.
Note: The experimenter may pause at this stage.
4. Image Processing and Analysis
- Open the spectral data (multistacked TIF) for the reference mirror and the brain tissue in ImageJ.
- Select the ROIs (regions of interest) for the opened image stacks—the central area for the reference mirror and the segment of an axon fiber for the brain tissue.
- Acquire the raw spectra for the selected ROIs by running Image → Stacks → Plot Z-axis profile on the ImageJ menu.
- Open the dark offset data, one taken for the reference mirror and the other taken for the brain tissue.
- Acquire the spectra for the dark offsets by running Image → Stacks → Plot Z-axis profile on the ImageJ menu.
- Save all the acquired spectra by using the Copy and Paste options.
Note: For SpeRe imaging, use of the central imaging field to minimize off-axis optical aberrations is recommended. The axon fibers may be structurally heterogeneous along their length. Hence, selecting the ROI on a small axon segment, typically < 5 µm, to minimize partial-volume artifact is recommended.
5. Baseline Correction and SpeRe Signal Analysis
- Subtract the offset spectra from the spectra of the reference mirror and the brain tissues.
- Normalize each spectrum by dividing the maximum intensity of the spectrum.
- Divide the normalized spectrum of the axon by the normalized spectrum of the reference mirror and subtract DC offset from the normalized spectrum.
- Measure the wavenumber frequency by fitting the acquired spectrum to a sinusoidal curve (see Table of Materials), and then convert the acquired wavenumber to periodicity by taking reciprocal.
- Convert the wavenumber periodicity to the axon diameter using the equation in Figure 4c.
Results
According to the protocol, a fixed brain slice was prepared with exogenous staining, a myelin-targeting fluorophore (see Table of Materials). SpeRe imaging was performed on the brain slice using a commercial hyperspectral confocal microscope in conjunction with confocal fluorescence imaging (Figure 4a). For SpeRe, input optical intensity was set to 5 µW/µm2 with a pixel dwell time of ~1 µs. This light dose is over an...
Discussion
SpeRe is a new label-free imaging modality based on spectral interferometry, which for the first time, offers the nanoscale information in live myelinated axons. In the current acquisition protocol, the spatial resolution for the axon diameter is of the order of 10 nm. Moreover, SpeRe utilizes orders-of-magnitude lower light dose compared to other super-resolution microscopies; thus, it is free from phototoxicity and photobleaching. SpeRe would provide a new avenue for studying nanoscale dynamics of the myelinated axons....
Disclosures
The authors declare competing financial interests: J. Kwon and M. Choi are inventors of the patent-pending technology described in this Article.
Acknowledgements
This work was supported by the Institute of Basic Science (IBS-R015-D1) and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1A6A1A03015642).
Materials
| Name | Company | Catalog Number | Comments |
| Glass cutter | - | - | Can be purchased in a local convenience store or online stores. |
| Nail polish | - | - | Can be purchased in a local convenience store or online stores. |
| Apochromat objective 40×, NA 1.1 | Leica Microsystems | 15506357 | Water-immersion type |
| Fluoromyelin Green | Thermo Fisher | F34651 | Alternatively, Fluoromyelin Red (F34652) can be used. |
| Leica SP8 TCS microscope | Leica Microsystems | SP8 | Refer to the "Configuration of microscope" in Introduction Section for details. |
| Imaging software | Leica Microsystems | LAS-X | - |
| Matlab | MathWorks | - | - |
| Mirror | Thorlabs | PF10-03-P01 | Coated with protected silver. |
| Phosphate-buffered saline (PBS) | Life technologies | 14190-136 | - |
| Paraformaldehyde | Biosolution | BP031a | 4% v/v in PBS |
| Cover slip | Thermo Fisher | 3306 | Thickness: #1 (0.13 to 0.17 mm) |
| Slide glass | Muto Pure Chemicals | 5116-20F | Thickness: ~1 mm |
| Super glue | Henkel | Loctite 406 | Use a dispensing equipment to avoid skin or eye contact. |
| Syringe pump | Brainetree Scientific | BS-8000 DUAL | - |
| Vibratome | Leica Biosystems | VT1200S | - |
| White-light laser | NKT photonics | EXB-6 | EXB-6 was discontinued and replaced by EXU-6. |
References
- Fernandez-Moran, H., Finean, J. B. Electron microscope and low-angle x-ray diffraction studies of the nerve myelin sheath. Journal of Cell Biology. 3, (1957).
- Blaurock, A. E. The spaces between membrane bilayers within PNS myelin as characterized by X-ray diffraction. Brain Research. 210, 383-387 (1981).
- Shim, S. -. H., et al. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proceedings of the National Academy of Sciences. , 13978-13983 (2012).
- Urban, N. T., Willig, K. I., Hell, S. W., Nägerl, U. V. STED nanoscopy of actin dynamics in synapses deep inside living brain slices. Biophysics Journal. 101, 1277-1284 (2011).
- Manley, S., et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nature Methods. 5, 155-157 (2008).
- Peters, A., Sethares, C. Is there remyelination during aging of the primate central nervous system?. Journal of Comparative Neurology. 460, 238-254 (2003).
- De Campos Vidal, B., Silveira Mello, M. L., Caseiro-Filho, A. C., Godo, C. Anisotropic properties of the myelin sheath. Acta Histochemica. 66, 32-39 (1980).
- Schain, A. J., Hill, R. A., Grutzendler, J. Label-free in vivo imaging of myelinated axons in health and disease with spectral confocal reflectance microscopy. Nature Medicine. 20, 443-449 (2014).
- Kwon, J., et al. Label-free nanoscale optical metrology on myelinated axons in vivo. Nature Communication. 8, 1832 (2017).
- Gage, G. J., Kipke, D. R., Shain, W. Whole Animal Perfusion Fixation for Rodents. JoVE. (65), e3564 (2012).
- Segev, A., Garcia-Oscos, F., Kourrich, S. Whole-cell Patch-clamp Recordings in Brain Slices. JoVE. (112), e54024 (2016).
- Waldchen, S., Lehmann, J., Klein, T., Van De Linde, S., Sauer, M. Light-induced cell damage in live-cell super-resolution microscopy. Scientific Reports. 5, 15348 (2015).
- Chang, J. B., et al. Iterative expansion microscopy. Nature Methods. 14, 593-599 (2017).
- Polishchuk, R. S., et al. Correlative light-electron microscopy reveals the tubular-saccular ultrastructure of carriers operating between Golgi apparatus and plasma membrane. Journal of Cell Biology. 148, 45-58 (2000).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved