A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Measuring the Spin-Lattice Relaxation Magnetic Field Dependence of Hyperpolarized [1-13C]pyruvate
In This Article
Summary
We present a protocol to measure the magnetic field dependence of the spin-lattice relaxation time of 13C-enriched compounds, hyperpolarized by means of dynamic nuclear polarization, using fast field-cycled relaxometry. Specifically, we have demonstrated this with [1-13C]pyruvate, but the protocol could be extended to other hyperpolarized substrates.
Abstract
The fundamental limit to in vivo imaging applications of hyperpolarized 13C-enriched compounds is their finite spin-lattice relaxation times. Various factors affect the relaxation rates, such as buffer composition, solution pH, temperature, and magnetic field. In this last regard, the spin-lattice relaxation time can be measured at clinical field strengths, but at lower fields, where these compounds are dispensed from the polarizer and transported to the MRI, the relaxation is even faster and difficult to measure. To have a better understanding of the amount of magnetization lost during transport, we used fast field-cycling relaxometry, with magnetic resonance detection of 13C nuclei at ~0.75 T, to measure the nuclear magnetic resonance dispersion of the spin-lattice relaxation time of hyperpolarized [1-13C]pyruvate. Dissolution dynamic nuclear polarization was used to produce hyperpolarized samples of pyruvate at a concentration of 80 mmol/L and physiological pH (~7.8). These solutions were rapidly transferred to a fast field-cycling relaxometer so that relaxation of the sample magnetization could be measured as a function of time using a calibrated small flip angle (3°-5°). To map the T1 dispersion of the C-1 of pyruvate, we recorded data for different relaxation fields ranging between 0.237 mT and 0.705 T. With this information, we determined an empirical equation to estimate the spin-lattice relaxation of the hyperpolarized substrate within the mentioned range of magnetic fields. These results can be used to predict the amount of magnetization lost during transport and to improve experimental designs to minimize signal loss.
Introduction
Magnetic resonance spectroscopic imaging (MRSI) can produce spatial maps of metabolites detected by spectroscopic imaging, but its practical use is often limited by its relatively low sensitivity. This low sensitivity of in vivo magnetic resonance imaging and spectroscopy methods stems from the small degree of nuclear magnetization achievable at body temperatures and reasonable magnetic field strengths. However, this limitation can be overcome by the use of dynamic nuclear polarization (DNP) to greatly enhance the in vitro magnetization of liquid substrates, which are subsequently injected to probe in vivo metabolism using MRSI1,2,3,4. DNP is capable of enhancing the magnetization of most nuclei with non-zero nuclear spin and has been used to increase in vivo MRSI sensitivity of 13C-enriched compounds such as pyruvate5,6, bicarbonate7,8, fumarate9, lactate10, glutamine11, and others by more than four orders of magnitude12. Its applications include imaging of vascular disease13,14,15, organ perfusion13,16,17,18, cancer detection1,19,20,21,22, tumor staging23,24, and quantification of therapeutic response2,6,23,24,25,26.
Slow spin-lattice relaxation is essential for in vivo detection with MRSI. Spin-lattice relaxation times (T1s) on the order of tens of seconds are possible for nuclei with low gyromagnetic ratios within small molecules in solution. Several physical factors influence the transfer of energy between a nuclear spin transition and its environment (lattice) leading to relaxation, including the magnetic field strength, temperature, and molecular conformation27. Dipolar relaxation is reduced in molecules for carbon positions with no protons directly attached, and deuteration of dissolution media can further reduce intermolecular dipolar relaxation. Unfortunately, deuterated solvents have limited abilities to extend in vivo relaxation. Increased relaxation of carbonyls or carboxylic acids (such as pyruvate) can occur at high magnetic field strengths due to chemical shift anisotropy. The presence of paramagnetic impurities from the fluid path during dissolution after polarization can cause rapid relaxation and need to be avoided or eliminated using chelators.
Very little data exist for the relaxation of 13C-containing compounds at low fields, where spin-lattice relaxation could be significantly faster. However, it is important to measure T1 at low fields to understand relaxation during preparation of the agent used for in vivo imaging, since the hyperpolarized contrast agents are usually dispensed from the DNP apparatus near or at the earth’s field. Additional physical factors such as 13C-enriched substrate concentration, solution pH, buffers and temperature also influence relaxation, and consequently have an effect on the formulation of the agent. All these factors are essential in the determination of key parameters in optimizing the DNP dissolution process, and the calculation of the magnitude of signal loss that occurs in transportation of the sample from the DNP apparatus to the imaging magnet.
Nuclear magnetic resonance dispersion (NMRD) measurements, i.e., T1 measurements, as a function of magnetic field are typically acquired using an NMR spectrometer. To acquire these measurements, a shuttling method could be used where the sample is first shuttled out of the spectrometer to relax at some field determined by its position in the fringe field of the magnet28,29,30 and then rapidly transferred back into the NMR magnet to measure its remaining magnetization. By repeating this process at the same point in the magnetic field but with increasing periods of relaxation, a relaxation curve can be obtained, which can then be analyzed to estimate T1.
We use an alternative technique known as fast field-cycling relaxometry31,32,33 to acquire our NMRD data. We have modified a commercial field-cycling relaxometer (see Table of Materials), for T1 measurements of solutions containing hyperpolarized 13C nuclei. Compared with the shuttle method, field-cycling enables this relaxometer to systematically acquire NMRD data over a smaller range of magnetic fields (0.25 mT to 1 T). This is accomplished by rapidly changing the magnetic field itself, not the sample location in the magnetic field. Therefore, a sample can be magnetized at a high field strength, "relaxed" at a lower field strength, and then measured by acquisition of a free-induction-decay at a fixed field (and Larmor frequency) to maximize signal. This means that the sample temperature can be controlled during the measurement, and the NMR probe does not need to be tuned at each relaxation field promoting automatic acquisition over the entire magnetic field range.
Focusing our efforts to the effects of dispensing and transporting the hyperpolarized solutions at low magnetic fields, this work presents a detailed methodology to measure the spin-lattice relaxation time of hyperpolarized 13C-pyruvate using fast field-cycling relaxometry for magnetic fields in the range of 0.237 mT to 0.705 T. The main results of using this methodology have been previously presented for [1-13C]pyruvate34 and 13C-enriched sodium and cesium bicarbonate35 where other factors such as radical concentration and dissolution pH have also been studied.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Sample Preparation
NOTE: Steps 1.1-1.8 are performed just once
- Prepare 1 mL of stock 13C-enriched pyruvic acid solution, widely used for in vivo research1,2,5,6, consisting of 15-mmol/L of triarylmethyl radical dissolved in [1-13C]pyruvic acid (see Table of Materials). Aliquots from this stock solution will be used for the samples that will be individually polarized and subsequently undergo relaxometry at different magnetic fields. A representation of the [1-13C]pyruvic acid molecule is shown in Figure 1.
- On the dynamic nuclear polarizer software interface (see Table of Materials), click on the Cooldown button to lower the temperature of the variable temperature insert (VTI) to 1.4 K.
- Once the DNP has reached the desired temperature, load 10 µL of the stock solution in a sample cup, open the turret doors and insert the cup into the VTI using an insertion wand specifically designed for this task.
- After that, quickly extract the wand and make sure the cup is released. Then close the turret doors and continue with the following steps while the temperature of the VTI goes back to 1.4 K.
- Prepare the DNP to run a microwave sweep in order to find the optimal RF frequency for hyperpolarization of the stock solution.
- On the computer controlling the spectrometer (part of the DNP system), establish the communication between the spectrometer and the DNP control software by double-clicking on HyperTerminal icon, previously configured with the appropriate serial communication parameters.
- Once the communication is established, launch the RINMR software, type in its command line .HYPERSENSENMR, and then press enter.
- After that, a new window will be shown on the screen and on it type the number one (1) in the Configuration Number field. Then, click on the Select Configuration button.
- Click on the button Do microwave sweep. A small window with a descending counter of seconds will be launched indicating that the spectrometer is ready and it will be waiting for periodic trigger signals, coming from the DNP control software, to sample the polarization.
- On the DNP control software, select the Calibrate tab and click on the Generate button.
- Using the calibration setup window, enter the following information: Start Frequency = 94.117 GHz, End Frequency = 94.137 GHz, Step Size = 1 MHz, Step Duration = 300 s, Power = 50 mW, Liquid Helium Level = 65%, and Temperature = 1.4 K.
- Click on the button Generate, which will close the setup window and return to the Calibrate tab that will display the number of steps and the time required to perform the desired microwave sweep.
- Once the desired VTI temperature is achieved, click the Enable button and then Start to initialize the microwave sweep process.
- At the end of the microwave sweep, recover the sample and record the optimal frequency where the maximum polarization is achieved. This optimal frequency is defined as the polarization frequency that provides the maximum polarization as shown in Figure 2. This frequency will be used for hyperpolarizing all the aliquots obtained from that specific stock solution of pyruvic acid.
- Prepare 250 mL of stock dissolution medium using a solution of 40-mmol/L Tris base, 50 mmol/L of sodium chloride, and 80-mmol/L sodium hydroxide in de-ionized water. Add ethylenediaminetetraacetic acid (EDTA) at a concentration of 100 mg/L to sequester any metal ion contamination. Similarly to the pyruvic acid stock solution, this dissolution medium will be used for all the different samples that will be polarized. Refer to the Table of Materials for more specific details regarding the chemicals used.
- Also, prepare 500 mL of stock cleaning solution consisting of 100 mg/L EDTA dissolved in deionized water. Approximately 10 mL of this cleaning solution is used after each polarization to clean the dissolution path of the DNP.
NOTE: Steps 1.9-1.27 are performed for each individual sample. - Cool the DNP apparatus to 1.4 K in preparation of hyperpolarizing a [1-13C]pyruvic acid sample by pressing the Cooldown button in the DNP main window.
- If the software used for the spectrometer is already active with configuration 1 selected, proceed with the following steps. Otherwise, perform steps 1.5.1 to 1.5.3 and then continue with the following steps.
- After verifying that configuration 1 is selected in the window controlling the DNP's spectrometer, click on the Solid Build Up button.
- Enter the file name SSBuilupXXX, where "XXX" is a number in the sequence of files stored with build-up data. This number is automatically incremented by the software. Then click OK. Similarly to the microwave sweep case, a small window with a descending counter of seconds will be launched indicating that the spectrometer is ready and it will be waiting for periodic trigger signals, coming from the DNP control software, to sample the polarization.
- Using the pyruvic acid - OX063 stock solution prepared in step 1.1, weigh out 30 mg in a sample cup.
- When the desired VTI temperature is achieved (1.4 K) click on Insert Sample, then select Normal Sample and then click on Next. Following the safety precautions displayed on the screen, insert the cup in the cold DNP apparatus, using a long wand specifically designed for this task.
- Once the cup is inserted, the wand removed, and the DNP doors closed, click Next and then Finish. At that point the hyperpolarizer system lowers the sample cup to the irradiation chamber partially filled (65%) with liquid helium.
- Wait until the temperature has returned to 1.4K and then click on the Polarize Sample button.
- In the new pop up window, set the frequency value to that obtained from the microwave sweep in step 1.6. In the same window, also set the power to 50 mW and the sampling time to 300 s. Click on Next, check the Enable Build-up Monitoring box, and then click on Finish.
NOTE: Once the polarization is started, the DNP control software generates trigger signals every 300 s to instruct the spectrometer to sample the polarization using a small tip angle. That way, the spectrometer software adds a sample point to a solid-state magnetization curve, now displayed in both the spectrometer software and in the DNP control software under the tab Polarization Build-Up. After the 4th sample and every sample after that, the spectrometer software fits the curve to an exponential growth function of the form:
S = A *exp(-t/Tp) + y0
where A is the polarization amplitude, in arbitrary units, t is the sampling time, Tp is the polarization time constant (both in seconds), and y0 is an offset. Based on the fitted parameters, the software also calculates the percentage polarization achieved up to that point in time, which is also displayed in the DNP's Polarization Status tab. - Polarize until the build-up of the solid-state magnetization reaches at least 95% of maximum (approximately one hour).
- While the sample is polarizing, prepare the Fast-Field-Cycling Relaxometer as explained in Section 2 below.
- When the desired polarization is achieved, click on Run Dissolution and under Method, select Pyruvic Acid Test. Then, click on Next.
- Following the instructions on the screen, open the DNP turret doors and load the heating and pressurizing chamber at the top of the apparatus with ~ 4.55 mL of the dissolution medium prepared in section 1.5 to produce a concentration of 80-mmol/L pyruvate upon dissolution at a pH of ~7.75 and temperature of ~37 °C.
- Position the recovering wand in the right position, close the turret doors, and at the computer click on Next and then on Finish. At that point the dissolution media will be superheated until the pressure reaches 10 bar.
- Once the 10 bar pressure is attained, the frozen and hyperpolarized pyruvate is automatically lifted from the liquid helium bath, quickly mixed, and thawed with the superheated dissolution media and ejected through a capillary tubing into a pear-shaped flask. While the hyperpolarized pyruvate/dissolution media mixture is ejected, constantly swirl the flask to ensure a homogeneous mixture.
- When all the mixture has been ejected, quickly draw 1.1 mL of the liquid into a syringe, transfer to a pre-warmed (37 °C) 10-mm-diameter NMR tube, and rapidly transport to the field-cycling relaxometer (See step 2.2.12).
- Dispense the remaining aliquot of every pyruvate dissolution into a 0.55-T benchtop NMR spectrometer (see Table of Materials) to check for possible systematic experimental effects.
- Immediately clean DNP fluid path using clean dissolution medium followed by ethanol. Blow helium gas through fluid path to remove remaining cleaning fluids and purge path of oxygen. Clean all glassware.
- After each measurement, record the pH of samples from both the bench top spectrometer and the field-cycling relaxometer.
NOTE: Each T1 measurement is a separate hyperpolarized dissolution from the DNP apparatus, so care is required to assure measurement-to-measurement reproducibility of the sample composition. This is accomplished by weighing all agents and solvents with a precision of 0.1 mg to assure accurate and reproducible preparation of the final hyperpolarized solutions.
2. Relaxometry
NOTE Please refer to Table 1 for a better understanding of the selection and use of the different parameters described in the following steps. Prior to dissolution, the relaxometer flip angle must be calculate and the relaxometer must be setup and ready for measurement of the hyperpolarized solution (see below).
- Flip-angle calibration
- Prepare 1 mL of neat [1-13C]pyruvic acid in an NMR tube and add a gadolinium contrast agent to reduce the T1 of the 13C nuclei to a value of less than 200 ms but more than 50 ms.
- Seal the NMR tube so it can be used multiple times as a calibration standard.
- Using the depth gauge of the relaxometer, set the depth of insertion of the NMR tube to the proper height to ensure the sample will be located at the center of the relaxometer RF coil.
- Mark the insertion depth of the 13C pyruvate calibration standard with adhesive tape to ensure repeatability.
- Place the depth stopper on the NMR tube to the position indicated by the tape and insert this calibration standard into the bore of the field-cycling relaxometer. Use a weight to keep the NMR tube in position.
- Open the instrument air valve and from the relaxometer front panel set the temperature controller to 37 °C. That will maintain the temperature of the sample at 37°C (± 0.5°C) using heated air during the experiment.
- Setup the field-cycling relaxometer hardware to acquire 13C nuclei signals. That includes installing and energizing the external shim coil (see Table of Materials), tuning and matching the RF coil to 8 MHz (~0.75 T for 13C nuclei), and using the appropriate λ/4 cable.
- In the instrument software, perform the following steps:
- Select the Main par tab
- Click on the cell next to the label Experiment and scroll down in the pop-up window to select the pulse sequence "13CANGLE.FFC ".
- Set the following acquisition parameters: RFA = 5; SWT = 0.005, RD = 0.5, BPOL = 30 MHz, TPOL = 0.5.
- Select the Acq. par tab and then select the Basic sub-tab.
- Click on the cell next to the label Nucleus and scroll down in the pop-up window to select 13C.
- Then, set the following parameters: SF = 8 MHz, SW = 1000000, BS = 652, FLTR = 100000, MS = 32.
- Select the Conf sub-tab.
- Set the following parameters: RINH = 25, ACQD = 25.
- Select the nDim sub-tab
- Set NBLK = 32, BINI = 2, BEND = 62.
- Select the Evaluation tab and then the Parameters sub-tab.
- Set the following parameters: EWIP = 10, EWEP = 128, EWIB = 1, EWEB = 32.
- Then, click the Start Acquisition icon to run the pulse sequence.
- Once the acquisition is finished, save the data, select the Evaluation dialog icon and from the analysis menu select WAM Window: Absolute Magnitude. Then select Report Sheet, Graphs and Export File and finally click on Execute.
- In the Report window find the RF pulse width that provides the maximum amplitude and fine-tune the value with the help of the cursor in the displayed graph, which is similar to the plots shown at the bottom row of Figure 3. This pulse width will be used for the parameter PW90 of the following experiments.
- Click the F1 icon to adjust the frequency shift of the relaxometer.
NOTE: WAM Window: Absolute Magnitude is a procedure to integrate the magnitude of a single or a sequence of free-induction decay acquisitions (FIDs) from the point defined by EWIP to the point specified by EWEP and from the block defined by EWIB to the block specified by EWEB.
- T1-Measurements
- Make sure the external shim coil is installed and energized.
- In the instrument software perform the following steps:
- Select the Main par tab
- Click on the cell next to the label Experiment and scroll down in the pop-up window to select the pulse sequence HPUB/S, which is shown in Figure 4.
- Set the following acquisition parameters: RFA = 25, T1MX = values between 3 and 5; SWT = 0.2, RD = 0, BRLX = Desired relaxation field in MHz (proton Larmor frequency).
- Select the Acq. par tab and then select the Basic sub-tab.
- Click on the cell next to the label Nucleus and scroll down in the pop-up window to select 13C.
- Then, set the following parameters: SF = 8 MHz, SW = 1000000, BS = 652, FLTR = 50000.
- Select the Conf sub-tab.
- Set the following parameters: PW90 equal to the value found in step 2.1.10, RINH = 25, ACQD = 25.
- Select the Puls sub-tab and set PW = 5.
- Select the nDim sub-tab and set NBLK = 100.
- Wait and get ready to receive the hyperpolarized solution to initiate the data acquisition.
- Immediately before inserting the sample into the relaxometer, manually start the pulse sequence from the console, to avoid inserting the sample into a null magnetic field. For this reason, it is important to ignore the first Free Induction Decay (FID) during the data analysis.
- Once the acquisition is done, save the data by clicking the Save button.
- Using the analysis software, integrate the magnitude of each FID signal to produce a data series comprised of sample magnetization as a function of time.
- Extract the spin-lattice relaxation time from a three-parameter exponential model using a standard non-linear least-squares fitting algorithm implemented in a commercial analytical software (see Table of Materials) assuming even weighting for all data:
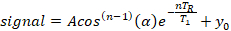
where A is the initial signal amplitude (y-intercept), T1 is the spin-lattice relaxation time, TR is the repetition time, which is a known value, y0 is the signal offset, and cos(n-1)(α) is a correction for loss of longitudinal magnetization at the nth measurement for a flip angle, α.
Access restricted. Please log in or start a trial to view this content.
Results
Figure 2 presents an example of a high-resolution full-range microwave sweep for pyruvic acid. For the presented case, that optimal microwave frequency corresponds to 94.128 GHz, highlighted in the figure insert. Our DNP system can normally work in the range of 93.750 GHz to 94.241 GHz with step size of 1 MHz, polarization time of up to 600 s, and power of up to 100 mW. A full range of frequencies is investigated only for novel substrates. However, based on previous experience with 13
Access restricted. Please log in or start a trial to view this content.
Discussion
The use of DNP to enhance signal acquisition is a technical solution to insufficient magnetic resonance signal available from 13C nuclei at limited concentrations, as those used in animal injections, but presents other experimental challenges. Each relaxation measurement shown in Figure 7 represents a measurement of a uniquely prepared sample because it cannot be repolarized after dissolution for remeasurement. This inevitably leads to experimental variability due to minor differe...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have no disclosures.
Acknowledgements
The authors would like to thank the Ontario Institute for Cancer Research, Imaging Translation Program and the Natural Sciences and Engineering Research Council of Canada for funding this research. We also like to acknowledge useful discussions with Albert Chen, GE Healthcare, Toronto, Canada, Gianni Ferrante, Stelar s.r.l., Italy, and William Mander, Oxford Instruments, UK.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| [1-13C]Pyruvic Acid | Sigma-Aldrich, St. Louis, MO, USA | 677175 | |
| 10mm NMR Tube | Norell, Inc., Morganton NC, USA | 1001-8 | |
| De-ionized water | |||
| Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA) | Sigma-Aldrich, St. Louis, MO, USA | E5134 | |
| HyperSense Dynamic Nuclear Polarizer | Oxford Instruments, Abingdon, UK | Includes the following: "DNP-NMR Polarizer" software used to control and monitor the whole DNP polarizer; "RINMR" used to monitor the solid state polarization levels; "HyperTerminal" used to communicate the DNP software with the RINMR software that monitors the solid state polarization level. Also includes the MQC bench top spectrometer to monitor the liquid state polarization in conjunction with it own RINMR software | |
| MATLAB R2017b | MathWorks, Natick, MA | Include scripts for non-linear fitting of magnetization decay over time and T1 NMRD analysis of hyperpolarized pyruvic acid. | |
| OX063 Triarylmethyl radical | Oxford Instruments, Abingdon, UK | ||
| pH meter - SympHony | VWR International, Mississauga, ON., Canada | SB70P | |
| ProHance | Bracco Diagnostics Inc. | Gadoteridol, Gd-HP-DO3A | |
| Pure Ethanol (100% pure) | Commercial Alcohols, Toronto, ON, Canada | P016EAAN | |
| Shim Coil | Developed in-house | ||
| Sodium Chloride | Sigma-Aldrich, St. Louis, MO, USA | S7653 | |
| Sodium Hydroxide | Sigma-Aldrich, St. Louis, MO, USA | S8045 | |
| SpinMaster FFC2000 1T C/DC | Stelar s.r.l., Mede (PV) Italy | Includes the software "AcqNMR" that is used to set experimental parameters, monitor the tuning and matching of the RF coil, loading different pulse sequences, calibrate flip angle, data acquisition and curve fitting, among other functions. Also includes a depth gauge, some weights and a depth stopper. | |
| Trizma Pre-Set Crystals (pH 7.6) | Sigma-Aldrich, St. Louis, MO, USA | T7943 |
References
- Golman, K., Zandt, R. I., Lerche, M., Pehrson, R., Ardenkjaer-Larsen, J. H. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Research. 66 (22), 10855-10860 (2006).
- Witney, T. H., Brindle, K. M. Imaging tumour cell metabolism using hyperpolarized 13C magnetic resonance spectroscopy. Biochemical Society Transactions. 38 (5), 1220-1224 (2010).
- Kurhanewicz, J., et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 13 (2), 81-97 (2011).
- Golman, K., et al. Cardiac metabolism measured noninvasively by hyperpolarized 13C MRI. Magnetic Resonance in Medicine. 59 (5), 1005-1013 (2008).
- Golman, K., in 't Zandt, R., Thaning, M. Real-time metabolic imaging. Proceedings of the National Academy of Science of the United States of America. 103 (30), 11270-11275 (2006).
- Day, S. E., et al. Detecting response of rat C6 glioma tumors to radiotherapy using hyperpolarized [1- 13C]pyruvate and 13C magnetic resonance spectroscopic imaging. Magnetic Resonance in Medicine. 65 (2), 557-563 (2011).
- Gallagher, F. A., et al. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature. 453 (7197), 940-943 (2008).
- Wilson, D. M., et al. Multi-compound polarization by DNP allows simultaneous assessment of multiple enzymatic activities in vivo. Journal of Magnetic Resonance. 205 (1), 141-147 (2010).
- Gallagher, F. A., et al. Production of hyperpolarized [1,4-13C2]malate from [1,4-13C2]fumarate is a marker of cell necrosis and treatment response in tumors. Proceedings of the National Academy of Science of the United States of America. 106 (47), 19801-19806 (2009).
- Chen, A. P., et al. Feasibility of using hyperpolarized [1-13C]lactate as a substrate for in vivo metabolic 13C MRSI studies. Magnetic Resonance Imaging. 26 (6), 721-726 (2008).
- Gallagher, F. A., Kettunen, M. I., Day, S. E., Lerche, M., Brindle, K. M. 13C MR spectroscopy measurements of glutaminase activity in human hepatocellular carcinoma cells using hyperpolarized 13C-labeled glutamine. Magnetic Resonance in Medicine. 60 (2), 253-257 (2008).
- Ardenkjaer-Larsen, J. H., et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 100 (18), 10158-10163 (2003).
- Ishii, M., et al. Hyperpolarized 13C MRI of the pulmonary vasculature and parenchyma. Magnetic Resonance in Medicine. 57 (3), 459-463 (2007).
- Lau, A. Z., Chen, A. P., Cunningham, C. H. Integrated Bloch-Siegert B(1) mapping and multislice imaging of hyperpolarized (1)(3)C pyruvate and bicarbonate in the heart. Magnetic Resonance in Medicine. 67 (1), 62-71 (2012).
- Lau, A. Z., et al. Rapid multislice imaging of hyperpolarized 13C pyruvate and bicarbonate in the heart. Magnetic Resonance in Medicine. 64 (5), 1323-1331 (2010).
- Golman, K., Ardenkjaer-Larsen, J. H., Petersson, J. S., Mansson, S., Leunbach, I. Molecular imaging with endogenous substances. Proceedings of the National Academy of Sciences of the United States of America. 100 (18), 10435-10439 (2003).
- Johansson, E., et al. Cerebral perfusion assessment by bolus tracking using hyperpolarized 13C. Magnetic Resonance in Medicine. 51 (3), 464-472 (2004).
- Johansson, E., et al. Perfusion assessment with bolus differentiation: a technique applicable to hyperpolarized tracers. Magnetic Resonance in Medicine. 52 (5), 1043-1051 (2004).
- Albers, M. J., et al. Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading. Cancer Research. 68 (20), 8607-8615 (2008).
- Chen, A. P., et al. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T-initial experience. Magnetic Resonance in Medicine. 58 (6), 1099-1106 (2007).
- Lupo, J. M., et al. Analysis of hyperpolarized dynamic 13C lactate imaging in a transgenic mouse model of prostate cancer. Magnetic Resonance Imaging. 28 (2), 153-162 (2010).
- von Morze, C., et al. Imaging of blood flow using hyperpolarized [(13)C]urea in preclinical cancer models. Journal of Magnetic Resonance Imaging. 33 (3), 692-697 (2011).
- Brindle, K. M., Bohndiek, S. E., Gallagher, F. A., Kettunen, M. I. Tumor imaging using hyperpolarized 13C magnetic resonance spectroscopy. Magnetic Resonance in Medicine. 66 (2), 505-519 (2011).
- Park, I., et al. Detection of early response to temozolomide treatment in brain tumors using hyperpolarized 13C MR metabolic imaging. Journal of Magnetic Resonance Imaging. 33 (6), 1284-1290 (2011).
- Bohndiek, S. E., et al. Detection of tumor response to a vascular disrupting agent by hyperpolarized 13C magnetic resonance spectroscopy. Molecular Cancer Therapeutics. 9 (12), 3278-3288 (2010).
- Witney, T. H., et al. Detecting treatment response in a model of human breast adenocarcinoma using hyperpolarised [1-13C]pyruvate and [1,4-13C2]fumarate. British Journal of Cancer. 103 (9), 1400-1406 (2010).
- Levitt, M. H. Spin dynamics: basics of nuclear magnetic resonance. , John Wiley & Sons. (2001).
- Mieville, P., Jannin, S., Bodenhausen, G. Relaxometry of insensitive nuclei: optimizing dissolution dynamic nuclear polarization. Journal of Magnetic Resonance. 210 (1), 137-140 (2011).
- Redfield, A. G. Shuttling device for high-resolution measurements of relaxation and related phenomena in solution at low field, using a shared commercial 500 MHz NMR instrument. Magnetic Resonance in Chemistry. 41 (10), 753-768 (2003).
- Grosse, S., Gubaydullin, F., Scheelken, H., Vieth, H. -M., Yurkovskaya, A. V. Field cycling by fast NMR probe transfer: Design and application in field-dependent CIDNP experiments. Applied Magnetic Resonance. 17 (2), 211-225 (1999).
- Kimmich, R., Anoardo, E. Field-cycling NMR relaxometry. Progress in Nuclear Magnetic Resonance Spectroscopy. 44 (3-4), 257-320 (2004).
- Guðjónsdóttir, M., Belton, P., Webb, G. Magnetic Resonance in Food Science: Challenges in a Changing World. , The Royal Society of Chemistry. 65-72 (2009).
- Anoardo, E., Galli, G., Ferrante, G. Fast-field-cycling NMR: Applications and instrumentation. Applied Magnetic Resonance. 20 (3), 365-404 (2001).
- Chattergoon, N., Martinez-Santiesteban, F., Handler, W. B., Ardenkjaer-Larsen, J. H., Scholl, T. J. Field dependence of T1 for hyperpolarized [1-13C]pyruvate. Contrast Media & Molecular Imaging. 8 (1), 57-62 (2013).
- Martínez-Santiesteban, F. M., Dang, T. P., Lim, H., Chen, A. P., Scholl, T. J. T1 nuclear magnetic relaxation dispersion of hyperpolarized sodium and cesium hydrogencarbonate-13C. NMR in Biomedicine. 30 (9), 3749(2017).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved