Method Article
Derivatization of Protein Crystals with I3C using Random Microseed Matrix Screening
In This Article
Summary
This article presents a method to generate protein crystals derivatized with I3C (5-amino-2,4,6-triiodoisophthalic acid) using microseeding to generate new crystallization conditions in sparse matrix screens. The trays can be set up using liquid dispensing robots or by hand.
Abstract
Protein structure elucidation using X-ray crystallography requires both high quality diffracting crystals and computational solution of the diffraction phase problem. Novel structures that lack a suitable homology model are often derivatized with heavy atoms to provide experimental phase information. The presented protocol efficiently generates derivatized protein crystals by combining random microseeding matrix screening with derivatization with a heavy atom molecule I3C (5-amino-2,4,6-triiodoisophthalic acid). By incorporating I3C into the crystal lattice, the diffraction phase problem can be efficiently solved using single wavelength anomalous dispersion (SAD) phasing. The equilateral triangle arrangement of iodine atoms in I3C allows for rapid validation of a correct anomalous substructure. This protocol will be useful to structural biologists who solve macromolecular structures using crystallography-based techniques with interest in experimental phasing.
Introduction
In the field of structural biology, X-ray crystallography is regarded as the gold standard technique to determine the atomic-resolution structures of macromolecules. It has been utilized extensively to understand the molecular basis of diseases, guide rational drug design projects and elucidate the catalytic mechanism of enzymes1,2. Although structural data provides a wealth of knowledge, the process of protein expression and purification, crystallization and structure determination can be extremely laborious. Several bottlenecks are commonly encountered that hinder the progress of these projects and this must be addressed to efficiently streamline the crystal structure determination pipeline.
Following recombinant expression and purification, preliminary conditions that are conducive to crystallization must be identified which is often an arduous and time-consuming aspect of X-ray crystallography. Commercial sparse matrix screens that consolidate known and published conditions have been developed to ease this bottleneck3,4. However, it is common to generate few hits from these initial screens despite using highly pure and concentrated protein samples. Observing clear drops indicates that the protein may not be reaching the levels of supersaturation required to nucleate a crystal. To encourage crystal nucleation and growth, seeds produced from pre-existing crystals can be added to the conditions and this allows for increased sampling of the crystallization space. Ireton and Stoddard first introduced the microseed matrix screening method5. Poor quality crystals were crushed to make a seed stock and then added systematically to crystallization conditions containing different salts to generate new diffraction-quality crystals that would not have otherwise formed. This technique was further improved by D'Arcy et al. who developed random microseed matrix screening (rMMS) in which seeds were introduced into a spare matrix crystallization screen6,7. This improved the quality of crystals and increased the number of crystallization hits on average by a factor of 7.
After crystals are successfully produced and an X-ray diffraction pattern is obtained, another bottleneck in the form of solving the 'phase problem' is encountered. During the data acquisition process, the intensity of diffraction (proportional to the square of the amplitude) is recorded but the phase information is lost, giving rise to the phase problem that halts immediate structure determination8. If the target protein shares high sequence identity to a protein with a previously determined structure, molecular replacement can be used to estimate the phase information9,10,11,12. Although this method is fast and inexpensive, model structures may not be available or suitable. The success of the homology model-based molecular replacement method drops significantly as sequence identity falls below 35%13. In the absence of a suitable homology model, ab initio methods, such as ARCIMBOLDO14,15 and AMPLE16, can be tested. These methods use computationally predicted models or fragments as starting points for molecular replacement. AMPLE, which uses predicted decoy models as starting points, struggles to solve structures of large (>100 residues) proteins and proteins containing predominately β-sheets. ARCIMBOLDO, which attempts to fit small fragments to extend into a larger structure, is limited to high resolution data (≤2 Å) and by the ability of algorithms to expand the fragments into a full structure.
If molecular replacement methods fails, direct methods such as isomorphous replacement17,18 and anomalous scattering at a single wavelength (SAD19) or multiple wavelengths (MAD20) must be used. This is often the case for truly novel structures, where the crystal must be formed or derivatized with a heavy atom. This can be achieved by soaking or co-crystallizing with a heavy atom compound, chemical modification (such as 5-bromouracil incorporation in RNA) or labelled protein expression (such as incorporating selenomethionine or selenocysteine amino acids into the primary structure)21,22. This further complicates the crystallization process and requires additional screening and optimization.
A new class of phasing compounds, including I3C (5-amino-2,4,6-triiodoisophthalic acid) and B3C (5-amino-2,4,6-tribromoisophthalic acid), offer exciting advantages over pre-existing phasing compounds23,24,25. Both I3C and B3C feature an aromatic ring scaffold with an alternating arrangement of anomalous scatters required for direct phasing methods and amino or carboxylate functional groups that interact specifically with the protein and provide binding site specificity. The subsequent equilateral triangular arrangement of heavy metal groups allows for simplified validation of the phasing substructure. At the time of writing, there are 26 I3C-bound structures in the Protein Data Bank (PDB), of which 20 were solved using SAD phasing26.
This protocol improves the efficacy of the structure determination pipeline by combining the methods of heavy metal derivatization and rMMS screening to simultaneously increase the number of crystallization hits and simplify the crystal derivatization process. We demonstrated this method was extremely effective with hen egg white lysozyme and a domain of a novel lysin protein from bacteriophage P6827. Structure solution using the highly automated Auto-Rickshaw structure determination pipeline is described, specifically tailored for the I3C phasing compound. There exists other automated pipelines that can be used such as AutoSol28, ELVES29 and CRANK230. Non-fully automated packages such as SHELXC/D/E can also be used31,32,33. This method is particularly beneficial to researchers who are studying proteins lacking homologous models in the PDB, by significantly reducing the number of screening and optimization steps. A prerequisite for this method is protein crystals or a crystalline precipitate of the target protein, obtained from previous crystallization trials.
Protocol
1. Experimental planning and considerations
- Use pre-existing crystals of the protein of interest, preferably generated through vapor diffusion crystallization. For a generalized protocol of vapor diffusion crystallization, see Benvenuti and Mangani34. Other methods of crystallization such as microbatch under oil and free interface diffusion will require harvesting the crystals prior to crushing to generate microseeds.
- In the preparation of a seed stock, use the highest quality crystals that can be sacrificed. The highest quality crystal can be judged visually based on morphology or the best diffracting crystal can be selected, if such data is available. It is very likely that even better quality crystals are obtained after optimization through seeding. In the case where no crystals are available, crystalline precipitate such as spherulites and needles can be used.
- Identify salt crystals. Salt crystals can grow in crystallization screens and can look like protein crystals. Using salt crystals in rMMS will provide no benefit and will waste precious sample, so it is important to eliminate salt false positives.
- Salt crystals are loud when they are crushed. Crystals must be crushed to generate a seed stock, so this strategy is particularly relevant. If an audible crack sound is heard when crushing up the crystals, the crystal is likely to be salt.
- If the protein contains tryptophan and tyrosine residues, use ultraviolet fluorescence microscopy to identify protein crystals which fluoresce under these lighting conditions.
- Use Izit dye (methylene blue) to stain protein crystals to differentiate them to salt crystals which remain relatively unstained. However, this procedure is more destructive and is only recommended if one has crystals to spare from replicates of the same drop.
NOTE: Although the aforementioned tests may give promising results, salt crystals may still be mistaken for protein crystals. In this case, diffraction experiments can be used to definitively discern between a protein and salt crystal.
2. Preparation of lithium I3C stock
- Measure out 120 mg of I3C (5-amino-2,4,6-triiodoisophthalic acid) into a 1.5 mL microcentrifuge tube.
- Dissolve I3C in 200 µL of 2 M lithium hydroxide. The solution can be gently heated using a heat block at 40-60 °C to encourage dissolution. The resulting lithium I3C solution should be brown and has a concentration of 1 M.
CAUTION: Lithium hydroxide is corrosive. Safety glasses, gloves and a lab coat should be worn. - Measure the pH of the solution. If necessary, add small amounts of 1 M hydrochloric acid or 2 M lithium hydroxide to adjust the pH to between 7 and 8. Add milliQ water to make the final solution volume to 400 µL. The concentration of the I3C stock solution is 0.5 M.
NOTE: Step 2.3 is optional. The pH of the solution should be between pH 7-8 prior to any pH adjustment. This step should be performed if the protein of interest is strongly affected by pH. The protocol can be paused here. Lithium I3C can be kept in the dark at 4 °C for at least two weeks35.
3. Addition of I3C to the protein stock
- Method 1
- Add stock lithium I3C to a 150 µL aliquot of the target protein. The final concentration should be between 5-40 mM lithium I3C.
- Method 2 (gentler method)
- Prepare a protein dilution buffer that matches the buffer of the target protein. To this dilution buffer, add stock lithium I3C to give a concentration of lithium I3C between 10-80 mM.
- Dilute the protein 1:1 with protein dilution buffer to give a final concentration of lithium I3C between 5-40 mM.
NOTE: Some proteins will precipitate upon coming into contact with high concentrations of lithium I3C in method 1, while other proteins can tolerate it. Method 2 reduces the likelihood of precipitation. However, this method halves the protein concentration. For proteins that do not have an established crystallization protocol, a protein concentration of 10 mg/mL is generally recommended for initial crystallization screening. An initial molar ratio of I3C to protein of 8 is recommended. Protein concentration and molar ratio of I3C to protein can be optimized after the initial screen.
4. Making a seed stock
- Make a rounded probe for crushing crystals.
- With a Bunsen burner on the blue flame, heat a Pasteur pipette towards its middle. Using a tweezer, pull the end of the Pasteur pipette to draw it out into a thin diameter of less than 0.3 mm.
- Once the midsection is thin enough, hold that segment in the flame to separate the pipette at this point and round the end of the pipette to finish the glass probe.
NOTE: Rounded probe crystal crushers are sold by third party vendors. These are an alternative to making rounded probes.
- Place five 1.5 mL microcentrifuge tubes on ice.
- Under a light microscope, examine the crystallization tray for a suitable condition to generate microcrystals. Ideally, good morphology large crystals are selected. However, this technique also works with poor morphology crystals, needles, plates, microcrystals and spherulites.
- Open up the crystallization tray well. For 96 well crystallization trays sealed with plastic, use a scalpel to cut the plastic sealing the well. For hanging drop trays sealed with grease, the coverslip can be removed using tweezers and inverted onto an even surface.
- Transfer 70 µL of reservoir solution to a microcentrifuge tube and chill it on ice. To the other microcentrifuge tubes, add 90 µL of reservoir solution and return to ice to chill.
NOTE: If the reservoir does not have enough volume or does not exist (in the case of microbatch under oil), create crystallization reservoir by mixing the appropriate reagents. - Agitate the crystal in the drop using the crystal probe to thoroughly crush it up. The crystal needs to be completely crushed up which can be monitored under the microscope.
- Remove all the liquid from the drop and transfer it to the microcentrifuge tube with the reservoir solution. Mix and subsequently take 2 µL of mixture from the microcentrifuge tube and add it back to the well. Rinse the well with the solution and transfer it to the microcentrifuge tube. Repeat this rinse step once more. From this point on, keep the microcentrifuge tube cold to avoid melting the microseeds in the mixture.
- Vortex the tube at maximum speed at 4 °C for 3 min, stopping regularly to chill the tube on ice to prevent overheating.
NOTE: Some microseeding protocols add a polytetrafluoroethylene seed bead to the microcentrifuge tube to aid crystal crushing7,36. We have employed the technique without the use of a seed bead with success, but see no problems with utilizing a seed bead to crush up crystals. - Make a 1 in 10 serial dilution of the seed stock by sequentially transferring 10 µL between the chilled reservoir solutions.
- Store seed stocks that will not be used immediately at -80 °C.
5. Setting up an rMMS screen
- Setting up a 96 well screening plate using a liquid dispensing robot. In the absence of a robot, a multichannel pipette may also be used.
- Transfer 75 µL from a deep well block to a 96 well crystallization tray. Add 1 µL to the crystallization drop and 74 µL to the reservoir.
- Transfer 1 µL of protein supplemented with lithium I3C, made in step 2, to the crystallization drop.
- Transfer 0.1 µL of seed stock to the crystallization drop.
- Seal the plate with clear sealing tape and incubate the plate at a constant temperature to allow crystal growth.
- Setting up a hanging drop screens
- Grease the edges of the hanging drop wells (hanging drop crystallization trays can be found in 24 and 48 well formats).
- Transfer 500 µL crystallization solution into reservoir.
- Near the center of a glass cover slide, place a 1 µL drop of protein supplemented with lithium I3C, made in step 2.
- Add 1 µL of the crystallization solution to the drop.
- Transfer 0.1 µL of seed stock to the crystallization drop.
- Invert the cover slide and seal the crystallization well by pushing the cover slide into the grease.
- Incubate the plate at a constant temperature to allow crystal growth.
NOTE: With new and untested seed stocks, it is recommended to use the most concentrated seed stock to maximize the chances of getting crystallization hits. Subsequent conditions can be set up with reduced seed concentration to optimize the number of crystals.
- Inspect crystal trays under a microscope regularly for crystal growth. If crystals are of sufficient quality, they can be harvested for data collection. Crystals can also be used to generate new seed stocks and new rMMS screens to allow for iterative optimization.
6. Data collection
- Harvest crystals using cryoloops, cryoprotect the crystals and flash cool them in liquid nitrogen. For additional information on flash cooling crystals, refer to Teng37 and Garman and Mitchell38.
- During the cryoprotection stage, if the crystal is passed through a new aqueous solution, I3C can be lost from the crystal due to it leeching into the cryoprotection solution. Use lithium I3C in the cryoprotection solution at a concentration that matches the crystallization condition to mitigate this.
- Crystals grown using this protocol have successfully been cryoprotected using Parabar 10312 oil based cryoprotectant (Hampton Research).
NOTE: The protocol can be paused here while crystals are stored in liquid nitrogen.
CAUTION: Liquid nitrogen can cause cold burns. Liquid nitrogen can also cause asphyxiation if used in enclosed spaces.
- Mount the crystal on the X-ray source goniometer and collect diffraction data using the protocol specific for the X-ray source.
- This technique relies on anomalous signal from iodine atoms in I3C. Thus, select the energy of the X-ray to maximize this signal.
- Set synchrotron X-ray sources with tunable energies as low as possible. For many macromolecular crystallography beamlines, the lowest configurable energy is 8000 to 8500 eV.
- Rotating anode X-ray sources cannot be tuned. Commonly used anode sources with copper have the Kα edge at 8046 eV, which provides a good anomalous signal for iodine (f" = 6.9 e). Anode sources with chromium have a Kα edge at 5415 eV, which provides a large anomalous signal for iodine (f" = 12.6 e).
- Radiation damage is a significant problem during data collection as it will degrade the anomalous signal39. Select the exposure time and attenuation of the beam to achieve the best diffraction while minimizing radiation dose.
NOTE: In a similar phasing compound with the iodine atoms replaced with bromine atoms, radiation damage has been shown to cause the radiolysis of the carbon bromine bond and a reduction in the occupancy of the bromine atoms24.- Use inverse beam SAD data collection as a collection strategy. The data is collected in wedges, with opposite wedges collected after each other. This allows Friedel pairs to be collected with an equivalent dose, resulting in an improved measurement of anomalous signal less affected by radiation damage. For example, an eight wedge strategy to collect 360° would involve collecting the data in the order of wedge 1 (0°-45°), wedge 2 (180°-225°), wedge 3 (46°-90°), wedge 4 (225°-270°), wedge 5 (90°-135°), wedge 6 (270°-315°), wedge 7 (135°-180°) and wedge 8 (315°-360°).
NOTE: Continuous rotation is an alternative collection strategy to that of inverse beam data collection. For a recent comparison of the collection strategies, see Garcie-Bonte & Katona40.
- Use inverse beam SAD data collection as a collection strategy. The data is collected in wedges, with opposite wedges collected after each other. This allows Friedel pairs to be collected with an equivalent dose, resulting in an improved measurement of anomalous signal less affected by radiation damage. For example, an eight wedge strategy to collect 360° would involve collecting the data in the order of wedge 1 (0°-45°), wedge 2 (180°-225°), wedge 3 (46°-90°), wedge 4 (225°-270°), wedge 5 (90°-135°), wedge 6 (270°-315°), wedge 7 (135°-180°) and wedge 8 (315°-360°).
7. Data processing and structure solution
- Perform data reduction on the diffraction data using XDS41, with the aim of maximizing the anomalous signal. Data reduction input parameters are specific to the dataset and may require some trial and error. Here are some recommendations to start.
- Set FRIEDEL'S LAW=FALSE. Execute CORRECT twice, setting STRICT_ABSORPTION_CORRECT = TRUE and STRICT_ABSORPTION_CORRECT = FALSE. One run can have a higher anomalous signal than the other. Compare the anomalous signals between the runs using the 'Anomal Corr' and 'SigAno' disciplines in the output. This provides an indicator of data quality.
- Run SHELXC on the XDS_ASCII.HKL file for a more accurate indication of anomalous signal. The 'Ranom' discipline will give an indication of the anomalous signal at different resolutions.
- Run POINTLESS42 and AIMLESS43 to scale the data. In AIMLESS, set the parameter ANOMALOUS ON. If the GUI is used, select the option Separate anomalous pairs for outlier rejection and merging statistics. Testing different resolution cutoffs may be required to maximize anomalous signal.
- Solve the protein structure using Auto-Rickshaw automated crystal structure determination pipeline44. Auto-Rickshaw will attempt to solve the phase problem and build the crystal structure of the protein automatically with protein modelling and refinement software.
- For proteins without a homology model template, run the SAD protocol of Auto-Rickshaw in Advanced Mode. Enter the required parameters.
- Select PROTEIN as the molecule type.
- Enter the data collection wavelength in angstroms (Å).
- Select "I" as substructure element to indicate iodine atoms was used.
- Select "i3c" as substructure type to indicate I3C was the phasing molecule.
- Select "sub_direct" as the substructure determination method. This method employs SHELXD32 to search for the substructure.
- Select "3" as the number of expected substructure per monomer.
- Enter "1" as the resolution cutoff of substructure search. This allows Auto-Rickshaw to automatically determine a suitable resolution cutoff.
- Enter the number of residues in a single monomer, spacegroup of the dataset, and number of molecules in the asymmetric unit based on the Matthews coefficient.
- Select the appropriate dissemination level of X-ray data that suits the needs. Selecting "AutoRickshaw developers" will allow Auto-Rickshaw developers to troubleshoot the run if problems arise.
- Input the anomalous data as an mtz file.
- Input the protein sequence as a seq, pir or txt file. A seq file can be generated in a text editor (such as Notepad++9 on Windows or nano in Linux). Create a new file, enter the primary sequence of the protein as one long line or separated by line breaks. Save the file with the .seq file extension.
- Enter an institutional email address.
- For proteins without a homology model template, run the SAD protocol of Auto-Rickshaw in Advanced Mode. Enter the required parameters.
- Results are delivered via a web-link sent to the email address provided.
NOTE: AutoRickshaw is an automated pipeline that invokes various crystallography software packages to solve an X-ray crystal structure32,33,45,46,47,48,49,50,51,52,53,54,55,56,57,58. If the Auto-Rickshaw run fails to solve the structure, other Auto-Rickshaw settings can be tested. The structure determination method can be changed to "sub_phassade" to use Phaser59 instead of SHELXD32. The number of expected substructure per monomer can be also increased or decreased. - During the experimental phasing of the crystal structure, Auto-Rickshaw will attempt to position heavy atoms in the unit cell, creating a substructure. The equilateral triangle arrangement of iodine atoms in I3C presents an efficient way of validating the substructure. If step 6.3 fails, validating the substructure could aid in troubleshooting structure solution.
- Download the list of heavy atom sites from the Auto-Rickshaw results page. It is a hyperlink called "heavy atom sites". This will download a text file with the heavy atom sites.
- Change the file extension of the file from .txt to .pdb.
- Open the PDB file in Coot60. Turn on symmetry to see other heavy atoms from neighboring asymmetric units.
- Measure the distances between the heavy atoms, including across asymmetric units. I3C will appear as an equilateral triangle with a side length of 6 angstroms. The presence of a triangle with these dimensions indicates the placements of those heavy atoms are correct.
Results
Incorporating I3C into rMMS can generate new conditions supporting derivatized crystal growth
The efficacy of simultaneous rMMS screening and I3C derivatization was demonstrated in two proteins, hen egg white lysozyme (HEWL, obtained as a lyophilized powder) and the putative Orf11 lysin N-terminal domain (Orf11 NTD) from bacteriophage P68. Each protein was screened against PEG/ION HT under four different conditions including: unseeded, seeded, unseeded with I3C and seeded with I3C (Figure 1). For both proteins, the sole addition of I3C did not increase the number of conditions conducive to crystallization. In the case of Orf11 NTD, only one suitable condition was identified with and without I3C (Figure 1B). When I3C was added to the HEWL screens, the number of hits was reduced from 31 to 26, highlighting the added complexities of crystallisation when introducing phasing compounds (Figure 1A). Consistent with other studies, adding seed to commercial sparse matrix screens to generate an rMMS screen significantly increased the number of possible crystallization conditions for both proteins, resulting in a 2.1 and 6 fold increase for HEWL and Orf11 NTD, respectively6,61 (Figure 1). Most importantly, simultaneous addition of I3C and seed increased the number of hits relative to an unseeded screen, demonstrating a 2.3 and 7 fold increase for HEWL and Orf11 NTD, respectively. Many of the crystals from rMMS in the presence of I3C show excellent crystal morphology (Figure 2).
Seeding allows careful control of crystal number in I3C rMMS screens
In microseeding experiments, the number of seeds introduced into a crystallization trial can be controlled by dilution of the seed stock and this allows for precise control of nucleation in the drop7,36. This often allows larger crystals to form since there is reduced competition of protein molecules at nucleation sites. This advantage also extends to the I3C-rMMS method and has been demonstrated successfully in both HEWL and Orf11 NTD. Recreation of a crystallization condition identified from the I3C-rMMS screen with a diluted seed stock yielded fewer but larger crystals (Figure 3).
SAD phasing can be used to solve the structures from crystals derived from rMMS I3C screen
Crystals grown using the diluted seed stock shown in Figure 3 were used to solve the structure of the proteins using SAD phasing using diffraction data from a single crystal (Figure 4). Data was collected on the Australian Synchrotron MX1 beamline62. Detailed data collection and structure solution details are described elsewhere27.
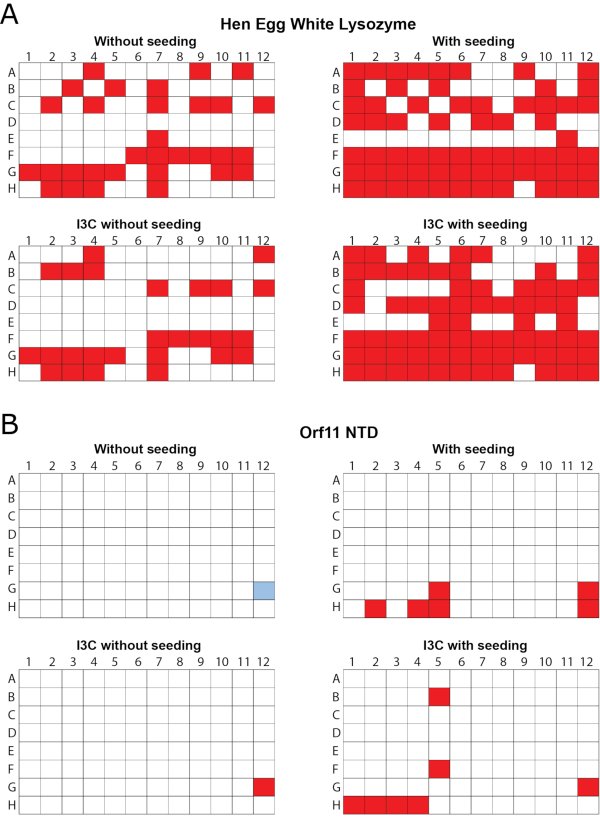
Figure 1 - rMMS was used to generate new conditions for crystal growth in the presence of I3C for two test proteins. 96 well vapor diffusion crystallization screens were carried out using commercial sparse matrix screens. (A) Hen egg white lysozyme was tested with the Index HT screen. Trays were seeded with HEWL crystals grown in 0.2 M ammonium tartrate dibasic pH 7.0, 20% (w/v) polyethylene glycol 3350. (B) Orf11 NTD from bacteriophage P68 was tested with the PEG/ION screen. Orf11 NTD trays were seeded from crystals from condition G12 from the unseeded screen, shown in blue. Conditions supporting crystal growth are shown in red. rMMS seeding in the presence and absence of I3C both gave significantly more crystal hits than unseeded trays. Figure adapted from Truong et al.27. Please click here to view a larger version of this figure.
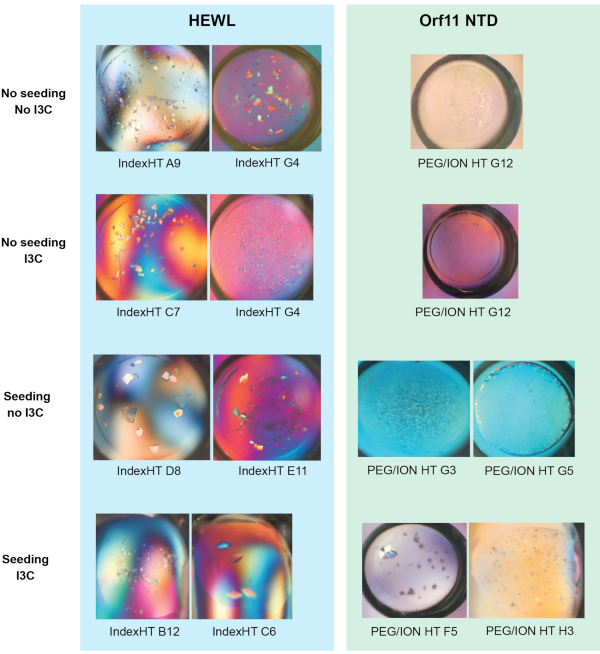
Figure 2 - Representative images of crystals grown from the vapor diffusion trials shown in Figure 1 (a) and (b). Figure adapted from Truong et al.27. Please click here to view a larger version of this figure.

Figure 3 - Dilution of the seed stock is an effective way to reduce nucleation in a crystallization condition found using the I3C-rMMS method, to control the number of crystals that form. Reducing nucleation within a drop often results in crystals growing to larger dimensions. Figure adapted from Truong et al.27. Please click here to view a larger version of this figure.

Figure 4 - Orf11 NTD (PDB ID 6O43) and HEWL (PDB ID 6PBB) were crystallized using the I3C-rMMS method and solved using Auto-Rickshaw SAD phasing. (A) Ribbon structures of HEWL and Orf11 NTD solved through experimental phasing. (B) I3C molecule bound to HEWL and Orf11 NTD. (C) Anomalous iodine atoms in I3C are arranged in an equilateral triangle of 6 Å. Thus the presence of this triangle in the phasing substructure indicates that there is an I3C molecule in that position. Please click here to view a larger version of this figure.
Discussion
Structure determination of a novel protein in the absence of a suitable homology model for molecular replacement requires experimental phasing. These methods require incorporation of heavy atoms into the protein crystal which adds a level of complexity to the structure determination pipeline and can introduce numerous obstacles that must be addressed. Heavy atoms can be incorporated directly into the protein through labelled expression using selenomethionine and selenocysteine. As this method is costly, laborious and can result in lower protein yields, labelled protein is often expressed after crystallization conditions has been found and optimized with unlabeled protein. Alternatively, crystals can be derivatized by soaking in a solution containing heavy atoms22,63,64. This method often uses high quality crystals and is therefore performed after a robust crystallization method has already been developed. Successfully obtaining a derivatized crystal using this method requires further optimization of soaking procedures and screening of different phasing compounds, therefore adding more time to an already laborious process.
Co-crystallization of the protein with the heavy atom can be performed at the screening stage, thus efficiently streamlining the process and reducing crystal manipulation steps that can cause damage. However, there still exists the potential scenario of obtaining few initial crystallization hits and the problem of choosing a compatible heavy atom compound. Many currently available phasing compounds are incompatible with precipitants, buffers and additives commonly found in crystallization conditions. They may be insoluble in sulphate and phosphate buffers, chelate to citrate and acetate, react unfavorably with HEPES and Tris buffers or become sequestered by DTT and β-mercaptoethanol21. As the I3C phasing compound does not suffer from these incompatibilities, it is a robust phasing compound that could be amenable to many different conditions.
In this study, a streamlined method of producing derivatized crystals ready for SAD phasing through simultaneous co-crystallization of the I3C phasing compound and rMMS is presented. The combination of both techniques increases the number of crystallization hits, with many of the conditions having improved morphology and diffraction characteristics. In both Orf11 NTD and HEWL test cases, new conditions in the I3C-rMMS screen were identified that were absent when I3C was not present. Potentially, I3C may bind favorably to the protein, facilitating the formation and stabilization of crystal contacts27. In turn, this may induce crystallization and possibly improve diffraction characteristics. Besides being a compound compatible with sparse matrix screens, I3C is also an attractive phasing compound due to its intrinsic properties. The functional groups that alternate with iodine on the aromatic ring scaffold allow specific binding to proteins. This leads to greater occupancy and potentially reduces background signal23. Furthermore, the arrangement of anomalous scatterers in an equilateral triangle is obvious in the substructure and can be used to rapidly validate binding of I3C (Figure 4B and 4C). Finally, it can produce an anomalous signal with tunable synchrotron radiation as well as chromium and copper rotating anode X-ray sources. Thus, it can be applied to many different workflows. As I3C is widely available and inexpensive to purchase, this approach is within reach for most structural biology laboratories.
There are several experimental considerations that must be addressed when using the I3C-rMMS method. This method cannot be applied if initial crystalline material of the protein cannot be obtained. In difficult cases, crystalline material from a homologous protein can also be used to generate seed stock. This cross-seeding approach to rMMS has shown some promising results7. Optimizing crystal number through dilution of the seed stock is a crucial step, which should not be overlooked, to maximize the chance of producing high quality large crystals and acquiring suitable diffraction data. If there are few I3C sites identified in the asymmetric unit, conditions conducive to crystallization should be further optimized with an increased concentration of I3C. This may increase the occupancy of I3C to maximize the anomalous signal and aid crystal derivatization.
There can be cases where this technique may not be the optimal method to derivatize protein crystals. As the size of a protein or protein-complex increases, the limited number of I3C sites on the protein surface may not provide sufficient phasing power to solve the structure. In these scenarios where protein size is suspected to be impeding phasing, selenomethionine labelling of the protein may be a more viable approach to phasing the protein. If the protein has adequate numbers of methionine residues in the protein (recommended having at least one methionine per 100 residues65) and high efficiency selenomethionine incorporation into a protein can be achieved (such as in bacterial expression systems66), multiple high occupancy selenium atoms will be present in the crystals to phase the structure.
In addition, some proteins may inherently be unsuited for derivatization with I3C. I3C binding sites on proteins are dependent on protein structure. There may exist proteins that naturally have few exposed patches compatible with I3C binding. Thus, it is not unforeseeable that there may be difficulties in co-crystallizing some target proteins with I3C.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was undertaken on the MX1 beamline at the Australian Synchrotron, part of ANSTO. The authors would like to acknowledge members of the Shearwin and Bruning laboratories for discussions on this work. The authors would also like to acknowledge Dr. Santosh Panjikar and Dr. Linda Whyatt-Shearwin who contributed to the original work that pioneered this protocol.
The following funding is acknowledged: Australian Research Council (grant Nos. DP150103009 and DP160101450 to Keith E. Shearwin); University of Adelaide (Australian Government Research Training Program stipend scholarship to Jia Quyen Truong and Stephanie Nguyen).
Materials
| Name | Company | Catalog Number | Comments |
| 10 mL disposable luer lock syringes | Adelab Scientific | T3SS10LAT | Used for dispensing vacuum grease for hanging drop crystal tray wells |
| 24 well tissue culture plate | Sigma Aldrich | CLS3527 | Used for hanging drop crystal tray |
| 3 inch wide Crystal Clear Sealing Tape | Hampton Research | HR4-506 | For 96 well crystallization screens set up by robot |
| 5-amino-2,4,6-triiodoisophthalic acid | Alfa Aesar | B22178 | Commonly referred to as I3C in the article |
| Art Robbins Intelli-Plate 96-2 Original | Hampton Research | HR3-297 | For 96 well crystallization screens set up by robot |
| Coverslips | Thermo Fisher Scientific | 18X18-2 | Coverslips for hanging drop crystal tray wells |
| Dow Corning vacuum grease | Hampton Research | HR3-510 | Used for sealing hanging drop crystal tray wells |
| Eppendorf Pipette 0.1 μL-2.5 μL | Eppendorf | 3120000011 | |
| Gilson Pipette 2 μL-20 μL | John Morris Group | 1153247 | |
| Gilson Pipette 20 μL-200 μL | John Morris Group | 1152006 | |
| Glass pasteur pipettes | Adelab Scientific | HIR92601.01 | |
| Hen Egg White Lysozyme | Sigma-Aldrich | L6876 | Approximately 95% pure |
| IndexHT screen | Hampton Research | HR2-134 | |
| Microscope illuminator | Meiji Techno | FT192/230 | Light source to illuminate crystallography experiments |
| PEG/ION HT screen | Hampton Research | HR2-139 | |
| Phoenix Liquid Dispenser | Art Robbins Instruments | 602-0001-10 | |
| Scalpel with scalpel blade no. 15 | Adelab Scientific | LV-SMSCPO15 | |
| Seed bead kit | Hampton Research | HR2-320 | Kit contains a glass probe for crushing crystals. A PTFE seed bead, designed for crushing crystals, is also part of the kit but not used in this protocol. |
| Stereo microscope | Meiji Techno | EMZ-5TR | Microscope for visualising crystallography experiments |
| Tweezers | Sigma-Aldrich | T5415 | |
| Vortex mixer | Adelab Scientific | RAVM1 |
References
- Zheng, H., Hou, J., Zimmerman, M. D., Wlodawer, A., Minor, W. The future of crystallography in drug discovery. Expert Opinion on Drug Discovery. 9 (2), 125-137 (2014).
- Oakley, A. J., Wilce, M. C. J. Macromolecular crystallography as a tool for investigating drug, enzyme and receptor interactions. Clinical and Experimental Pharmacology and Physiology. 27 (3), 145-151 (2000).
- Jancarik, J., Kim, S. H. Sparse matrix sampling. A screening method for crystallization of proteins. Journal of Applied Crystallography. 24, 409-411 (1991).
- Newman, J., et al. Towards rationalization of crystallization screening for small- To medium-sized academic laboratories: The PACT/JCSG+ strategy. Acta Crystallographica Section D: Biological Crystallography. 61 (10), 1426-1431 (2005).
- Ireton, G. C., Stoddard, B. L. Microseed matrix screening to improve crystals of yeast cytosine deaminase. Acta Crystallographica Section D: Biological Crystallography. 60 (3), 601-605 (2004).
- D'Arcy, A., Villard, F., Marsh, M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallographica Section D: Biological Crystallography. 63 (4), 550-554 (2007).
- D'Arcy, A., Bergfors, T., Cowan-Jacob, S. W., Marsh, M. Microseed matrix screening for optimization in protein crystallization: What have we learned. Acta Crystallographica Section:F Structural Biology Communications. 70 (9), 1117-1126 (2014).
- Taylor, G. The phase problem. Acta Crystallographica - Section D Biological Crystallography. 59 (11), 1881-1890 (2003).
- Rossmann, M. G. The molecular replacement method. Acta Crystallographica Section A. 46 (2), 73-82 (1990).
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., Read, R. J. Phaser crystallographic software. Journal of Applied Crystallography. 40 (4), 658-674 (2007).
- Millán, C., Jiménez, E., Schuster, A., Diederichs, K., Usón, I. ALIXE: a phase-combination tool for fragment-based molecular replacement. Acta Crystallographica Section D. 76 (3), 209-220 (2020).
- Liebschner, D., et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallographica Section D: Structural Biology. 75, 861-877 (2019).
- Abergel, C. Molecular replacement: Tricks and treats. Acta Crystallographica Section D: Biological Crystallography. 69 (11), 2167-2173 (2013).
- Pröpper, K., et al. Structure solution of DNA-binding proteins and complexes with ARCIMBOLDO libraries. Acta Crystallographica Section D: Biological Crystallography. 70 (6), 1743-1757 (2014).
- Rodríguez, D. D., et al. Crystallographic ab initio protein structure solution below atomic resolution. Nature Methods. 6 (9), 651-653 (2009).
- Bibby, J., Keegan, R. M., Mayans, O., Winn, M. D., Rigden, D. J. AMPLE: A cluster-and-truncate approach to solve the crystal structures of small proteins using rapidly computed ab initio models. Acta Crystallographica Section D: Biological Crystallography. 68 (12), 1622-1631 (2012).
- Green, D. W., Ingram, V. M., Perutz, M. F. The structure of haemoglobin - IV. Sign determination by the isomorphous replacement method. Proceedings of the Royal Society of London. Series A. Mathematical and Physical Sciences. 225 (1162), 287-307 (1954).
- Blow, D. M., Rossmann, M. G. The single isomorphous replacement method. Acta Crystallographica. 14 (11), 1195-1202 (1961).
- Wang, B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods in Enzymology. 115, 90-112 (1985).
- Hendrickson, W. A. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 254 (5028), 51-58 (1991).
- Pike, A. C. W., Garman, E. F., Krojer, T., Von Delft, F., Carpenter, E. P. An overview of heavy-atom derivatization of protein crystals. Acta Crystallographica Section D: Structural Biology. 72 (3), 303-318 (2016).
- Dauter, Z., Dauter, M., Rajashankar, K. R. Novel approach to phasing proteins: Derivatization by short cryo-soaking with halides. Acta Crystallographica Section D: Biological Crystallography. 56 (2), 232-237 (2000).
- Beck, T., Krasauskas, A., Gruene, T., Sheldrick, G. M. A magic triangle for experimental phasing of macromolecules. Acta Crystallographica Section D: Biological Crystallography. 64 (11), 1179-1182 (2008).
- Beck, T., Gruene, T., Sheldrick, G. M. The magic triangle goes MAD: Experimental phasing with a bromine derivative. Acta Crystallographica Section D: Biological Crystallography. 66 (4), 374-380 (2010).
- Beck, T., Da Cunha, C. E., Sheldrick, G. M. How to get the magic triangle and the MAD triangle into your protein crystal. Acta Crystallographica Section F: Structural Biology and Crystallization Communications. 65 (10), 1068-1070 (2009).
- Berman, H. M., et al. The Protein Data Bank. Nucleic Acids Research. , (2000).
- Truong, J. Q., Panjikar, S., Shearwin-Whyatt, L., Bruning, J. B., Shearwin, K. E. Combining random microseed matrix screening and the magic triangle for the efficient structure solution of a potential lysin from bacteriophage P68. Acta Crystallographica Section D: Structural Biology. 75 (7), 670-681 (2019).
- Terwilliger, T. C., et al. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallographica Section D: Biological Crystallography. 65 (6), 582-601 (2009).
- Holton, J., Alber, T. Automated protein crystal structure determination using ELVES. Proceedings of the National Academy of Sciences of the United States of America. 101 (6), 1537-1542 (2004).
- Skubák, P., Pannu, N. S. Automatic protein structure solution from weak X-ray data. Nature Communications. 4, (2013).
- Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallographica Section C: Structural Chemistry. 71, 3-8 (2015).
- Schneider, T. R., Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallographica Section D: Biological Crystallography. 58 (10), 1772-1779 (2002).
- Sheldrick, G. M. Macromolecular phasing with SHELXE. Zeitschrift fur Kristallographie. 217 (12), 644-650 (2002).
- Benvenuti, M., Mangani, S. Crystallization of soluble proteins in vapor diffusion for x-ray crystallography. Nature Protocols. , (2007).
- Beck, T. . Sticky triangles New tools for experimental phasing of biological macromolecules. , (2010).
- Luft, J. R., DeTitta, G. T. A method to produce microseed stock for use in the crystallization of biological macromolecules. Acta Crystallographica Section D: Biological Crystallography. 55 (5), 988-993 (1999).
- Teng, T. -. Y. Mounting of crystals for macromolecular crystallography in a free-standing thin film. Journal of Applied Crystallography. 23 (5), 387-391 (1990).
- Garman, E. F., Mitchell, E. P. Glycerol concentrations required for cryoprotection of 50 typical protein crystallization solutions. Journal of Applied Crystallography. 29, 584-587 (1996).
- Garman, E. F., Weik, M. X-ray radiation damage to biological samples: recent progress. Journal of Synchrotron Radiation. 26 (4), 907-911 (2019).
- Garcia-Bonete, M. J., Katona, G. Bayesian machine learning improves single-wavelength anomalous diffraction phasing. Acta Crystallographica Section A: Foundations and Advances. 75, 851-860 (2019).
- Kabsch, W. XDS. Acta Crystallographica Section D Biological Crystallography. 66 (2), 125-132 (2010).
- Evans, P. Scaling and assessment of data quality. Acta Crystallographica Section D: Biological Crystallography. 62 (1), 72-82 (2006).
- Evans, P. R., Murshudov, G. N. How good are my data and what is the resolution. Acta Crystallographica Section D: Biological Crystallography. 69 (7), 1204-1214 (2013).
- Panjikar, S., Parthasarathy, V., Lamzin, V. S., Weiss, M. S., Tucker, P. A. Auto-Rickshaw: An automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallographica Section D: Biological Crystallography. 61 (4), 449-457 (2005).
- Jones, T. A., Thirup, S. Using known substructures in protein model building and crystallography. The EMBO journal. 5 (4), 819-822 (1986).
- Kleywegt, G. J., Jones, T. A. Template convolution to enhance or detect structural features in macromolecular electron-density maps. Acta Crystallographica Section D: Biological Crystallography. 53 (2), 179-185 (1997).
- Perrakis, A., Morris, R., Lamzin, V. S. Automated protein model building combined with iterative structure refinement. Nature Structural Biology. 6 (5), 458-463 (1999).
- Morris, R. J., et al. Breaking good resolutions with ARP/wARP. Journal of Synchrotron Radiation. 11 (1), 56-59 (2004).
- Yao, D. Q., et al. SAD phasing by OASIS-2004: Case studies of dual-space fragment extension. Acta Crystallographica Section D: Biological Crystallography. 62 (8), 883-890 (2006).
- Hao, Q. ABS: A program to determine absolute configuration and evaluate anomalous scatterer substructure. Journal of Applied Crystallography. 37 (3), 498-499 (2004).
- Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallographica Section D: Biological Crystallography. 50 (5), 760-763 (1994).
- Sheldrick, G. M., Hauptman, H. A., Weeks, C. M., Miller, R., Usón, I. Ab initio phasing. International Tables for Crystallography. , 333-345 (2006).
- Smith, G. D. Matching selenium-atom peak positions with a different hand or origin. Journal of Applied Crystallography. 35 (3), 368-370 (2002).
- Pannu, N. S., McCoy, A. J., Read, R. J. Application of the complex multivariate normal distribution to crystallographic methods with insights into multiple isomorphous replacement phasing. Acta Crystallographica - Section D Biological Crystallography. 59 (10), 1801-1808 (2003).
- Pannu, N. S., Read, R. J. The application of multivariate statistical techniques improves single-wavelength anomalous diffraction phasing. Acta Crystallographica Section D: Biological Crystallography. 60 (1), 22-27 (2004).
- De La Fortelle, E., Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods in Enzymology. 276, 472-494 (1997).
- Cowtan, K. Joint CCP4 and ESF-EACBM Newsletter on Protein. Crystallography. 31, 34-38 (1994).
- Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallographica Section D: Biological Crystallography. 56 (8), 965-972 (2000).
- Read, R. J., McCoy, A. J. Maximum-likelihood determination of anomalous substructures. Acta Crystallographica Section D: Structural Biology. , (2018).
- Emsley, P., Lohkamp, B., Scott, W. G., Cowtan, K. Features and development of Coot. Acta Crystallographica Section D: Biological Crystallography. 66 (4), 486-501 (2010).
- Till, M., et al. Improving the success rate of protein crystallization by random microseed matrix screening. Journal of visualized experiments JoVE. (78), e50548 (2013).
- McPhillips, T. M., et al. Blu-Ice and the distributed control system: Software for data acquisition and instrument control at macromolecular crystallography beamlines. Journal of Synchrotron Radiation. 9 (6), 401-406 (2002).
- Nagem, R. A. P., Polikarpov, I., Dauter, Z. Phasing on Rapidly Soaked Ions. Methods in Enzymology. 374, 120-137 (2003).
- Taylor, G. L. Introduction to phasing. Acta Crystallographica Section D: Biological Crystallography. 66 (4), 325-338 (2010).
- Hendrickson, W. A., Ogata, C. M. Phase determination from multiwavelength anomalous diffraction measurements. Methods in Enzymology. 276, 494-523 (1997).
- Doublié, S. Production of Selenomethionyl Proteins in Prokaryotic and Eukaryotic Expression Systems. Macromolecular Crystallography Protocols. Methods in Molecular Biology. , 91-108 (2007).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved