A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Organoid-Derived Epithelial Monolayer: A Clinically Relevant In Vitro Model for Intestinal Barrier Function
In This Article
Summary
Here, we describe the preparation of human organoid-derived intestinal epithelial monolayers for studying intestinal barrier function, permeability, and transport. As organoids represent original epithelial tissue response to external stimuli, these models combine the advantages of expandability of cell lines and the relevance and complexity of primary tissue.
Abstract
In the past, intestinal epithelial model systems were limited to transformed cell lines and primary tissue. These model systems have inherent limitations as the former do not faithfully represent original tissue physiology, and the availability of the latter is limited. Hence, their application hampers fundamental and drug development research. Adult stem-cell-based organoids (henceforth referred to as organoids) are miniatures of normal or diseased epithelial tissue from which they are derived. They can be established very efficiently from different gastrointestinal (GI) tract regions, have long-term expandability, and simulate tissue- and patient-specific responses to treatments in vitro. Here, the establishment of intestinal organoid-derived epithelial monolayers has been demonstrated along with methods to measure epithelial barrier integrity, permeability and transport, antimicrobial protein secretion, as well as histology. Moreover, intestinal organoid-derived monolayers can be enriched with proliferating stem and transit-amplifying cells as well as with key differentiated epithelial cells. Therefore, they represent a model system that can be tailored to study the effects of compounds on target cells and their mode of action. Although organoid cultures are technically more demanding than cell lines, once established, they can reduce failures in the later stages of drug development as they truly represent in vivo epithelium complexity and interpatient heterogeneity.
Introduction
The intestinal epithelium acts as a physical barrier between the luminal content of the intestines and the underlying tissue. This barrier comprises a single epithelial layer of mainly absorptive enterocytes that are connected by tight junctions, which establish strong intercellular connections between adjacent cells. These cells form a polarized epithelial lining that separates the apical (lumen) and basolateral sides of the intestine, while simultaneously regulating paracellular transport of digested nutrients and metabolites. In addition to enterocytes, other important epithelial cells such as goblet, Paneth, and enteroendocrine cells also contribute to intestinal homeostasis by producing mucus, antimicrobial peptides, and hormones, respectively. The intestinal epithelium is constantly replenished by dividing leucine-rich repeat-containing G-protein-coupled receptor 5-positive (LGR5+) stem cells in the bottom of intestinal crypts producing transit-amplifying (TA) cells that migrate upwards and differentiate into other cell types1. Disruption of intestinal epithelial homeostasis by genetic and environmental factors, such as exposure to food allergens, medicinal compounds, and microbial pathogens, leads to disruption of intestinal barrier function. These conditions cause several intestinal diseases including inflammatory bowel disease (IBD), celiac disease, and drug-induced GI toxicity2.
Studies on the intestinal epithelium are performed using several in vitro platform systems such as membrane inserts, organs-on-a-chip systems, Ussing chambers, and intestinal rings.These platforms are suitable for establishing polarized epithelial monolayers with access to both apical and basolateral sides of the membrane, using transformed cell lines or primary tissue as models. Although transformed cell lines, such as the colorectal (adeno)carcinoma cell lines Caco-2, T84, and HT-29, are able to differentiate into polarized intestinal enterocytes or mucus-producing cells to some extent, they are not representative of the in vivo epithelium as several cell types are missing, and various receptors and transporters are aberrantly expressed3. In addition, as cell lines are derived from a single donor, they do not represent interpatient heterogeneity and suffer from reduced complexity and physiological relevance. Although primary tissues used in Ussing chambers and as intestinal rings are more representative of the in vivo situation, their limited availability, short-term viability, and lack of expandability make them unsuitable as a medium for high-throughput (HT) studies.
Organoids are in vitro epithelial cultures established from different organs such as the intestine, kidney, liver, pancreas, and lung. They are proven to have long-term, stable expandability as well as genetic and phenotypic stability and therefore are representative biological miniatures of the epithelium of the original organ with faithful responses to external stimuli4,5,6,7,8,9. Organoids are efficiently established from either resected or biopsied normal, diseased, inflamed, or cancerous tissue, representing heterogeneous patient-specific responses10,11,12,13,14,15,16. This paper demonstrates how to establish intestinal epithelial monolayers derived from organoid cultures. Monolayers have been successfully established from small intestinal as well as colonic and rectal organoid cultures. This model creates an opportunity to study the transport and permeability of the epithelial cells to drugs as well as their toxicological effects on the epithelium. Moreover, the model allows co-culture with immune cells and bacteria to study their interactions with the intestinal epithelium17,18,19. Furthermore, this model can be used to study responses to therapies in a patient-specific manner and initiate screening efforts to look for the next wave of epithelial barrier-focused therapeutics. Such an approach could be extended to the clinic and pave the way toward personalized treatments.
Although the epithelial monolayers in this protocol are prepared from human normal intestinal organoids, the protocol can be applied and optimized for other organoid models. Epithelial organoid monolayers are cultured in intestinal organoid expansion medium containing Wnt to support stem cell proliferation and represent intestinal crypt cellular composition. Intestinal organoids can be enriched to have different intestinal epithelial fates, such as enterocytes, Paneth, goblet, and enteroendocrine cells, by modulating Wnt, Notch, and epidermal growth factor (EGF) pathways. Here, after the establishment of monolayers in expansion medium, they are driven toward more differentiated intestinal epithelial cells, as described previously20,21,22,23,24,25. For screening purposes, depending on the mode of action of the compound of interest, its target cells, and the experimental conditions, the monolayers can be driven toward the cellular composition of choice to measure the effects of the compound with relevant functional readouts.
Protocol
1. Preparing reagents for culture
NOTE: Perform all steps inside a biosafety cabinet and follow standard guidelines for working with cell cultures. Ultraviolet light is used for 10 min before starting up the biosafety cabinet. Before and after use, the surface of the biosafety cabinet is cleaned with a tissue paper drenched in 70% ethanol. To facilitate the formation of three-dimensional drops of extracellular matrix (ECM), keep a prewarmed stock of 96-, 24-, and 6-well plates ready in the incubator at 37 °C.
- Basal medium preparation
- Prepare basal medium (BM) in a 500 mL Advanced Dulbecco's Modified Eagle Medium with Ham's Nutrient Mixture F-12 (Ad-DF) medium bottle by adding 5 mL of 200 mM glutamine, 5 mL of 1 M 4-(2-hydroxyethil)-1piperazineethanesulfonic acid (HEPES), and 5 mL of penicillin/streptomycin (pen/strep) solutions (10,000 U/mL or 10,000 µg/mL). This can be stored in the refrigerator at 4 °C for at least 4 weeks.
- Wnt sources
- Prepare Wnt3a-conditioned medium (Wnt3aCM) according to the previously described method26.
NOTE: Recently, a next-generation surrogate Wnt (NGS-Wnt), which also supports expansion of human intestinal organoids, has been generated27.
- Prepare Wnt3a-conditioned medium (Wnt3aCM) according to the previously described method26.
- Intestinal organoid base medium preparation
NOTE: Use all growth factors and reagents according to the manufacturer's recommendations. Use small aliquots and avoid freeze-thaw cycles; functional growth factors are essential for successful organoid culture.- Prepare concentrated 2x intestinal organoid base medium (2x IBM) by supplementing BM with 1 µM A83-01, 2.5 mM N-acetylcysteine, 2x B27 supplement, 100 ng/mL human epidermal growth factor (hEGF), 10 nM gastrin, 200 ng/mL hNoggin, and 100 µg/mL of an antimicrobial formulation for primary cells (see the Table of Materials).
- Aliquot the 2x IBM and freeze at -20 °C for up to 4 months. When needed, thaw an aliquot overnight at 4 °C or for several hours at room temperature (RT).
- To prepare intestinal organoid expansion medium (IEM), supplement 2x IBM with either 50% Wnt3aCM or 50% BM and 0.5 nM NGS-Wnt, 250 ng/mL human Rspondin-3 (hRspo3), 10 mM nicotinamide, and 10 µM SB202190.
- Intestinal organoid differentiation medium preparation
- Prepare enterocyte differentiation medium (eDM) by supplementing 2x IBM with 50% BM, 250 ng/mL hRspo3, and 1.5 µM Wnt pathway inhibitor (IWP-2). Store eDM at 4 °C for up to 10 days.
- Prepare combination differentiation medium (cDM) by supplementing 2x IBM with either 40% BM and 10% Wnt3aCM or 50% BM and 0.1 nM NGS-Wnt, 250 ng/mL hRspo3, 10 µM DAPT and 100 nM PD0325901. Store cDM at 4 °C for up to 10 days.
- Manipulation of extracellular matrix (ECM)
NOTE: Prepare the extracellular matrix (ECM) (see the Table of Materials) according to the manufacturer's recommendation.- Thaw ECM overnight on ice; transfer the ECM from the bottle to a 15 mL conical tube using a 5 mL pipette, both pre-cooled at -20 °C. Refreeze aliquots only once at -20 °C. Once thawed, store the ECM in a refrigerator at 4 °C for up to 7 days. Incubate for at least 30 min on ice before use.
NOTE: Mix ECM properly and ensure that it is cold before embedding crypts or organoids.
- Thaw ECM overnight on ice; transfer the ECM from the bottle to a 15 mL conical tube using a 5 mL pipette, both pre-cooled at -20 °C. Refreeze aliquots only once at -20 °C. Once thawed, store the ECM in a refrigerator at 4 °C for up to 7 days. Incubate for at least 30 min on ice before use.
2. Organoid cultures
- Establishing cultures from frozen organoids
NOTE: Let BM reach RT, and keep a 12 mL aliquot, warmed to 37 °C, ready before starting the procedure of thawing one cryovial containing frozen organoids.- Thaw the organoid cryovial rapidly by agitating in a 37 °C water bath until only a sliver of ice remains. Immediately add 500 µL of warm BM dropwise to the cryovial, and pipet up and down a few times to dilute the freezing medium and mix the contents carefully.
- Using a P1000 pipette, transfer the organoids to a 15 mL conical tube, and add another 1 mL of warm BM dropwise while gently mixing the bottom of the tube. Pipet up and down a few times to dilute the freezing medium and mix the contents carefully.
- Add up to 12 mL of warm BM dropwise to the 15 mL conical tube containing the organoids, and pipet up and down with a 10 mL sterile pipette to gently resuspend the organoids.
- Centrifuge the organoid suspension for 5 min at 85 × g and 8 °C. Discard the supernatant carefully without disturbing the pellet, and resuspend the organoids in 30% v/v of IEM supplemented with 10 µM Y27632 or other rho-associated coiled-coil-forming protein serine/threonine kinase inhibitor (ROCK inhibitor). Place the tube on ice.
- Add 70% v/v of ECM in the 15 mL conical tube containing the organoids. Mix the organoid suspension keeping the 15 mL conical tube on ice, and seed 5 µL of the suspension to check the density (Figure 1A). Continue plating if the density is appropriate; if the density is too high, add more IEM/ECM solution in the same ratio of 30-70% v/v, respectively.
- In each well of a prewarmed 24-well plate, seed 50 µL of the organoid suspension by pipetting 5 separate drops of 10 µL (Figure 1B). Turn the plate upside down, and leave it in the biosafety cabinet for 5 min. Transfer the plate still upside down to the 37 °C incubator, and leave it for another 30 min.
- Add 500 µL of IEM with 10 µM ROCK inhibitor to each well, and transfer the plate to the incubator. Image one drop regularly to monitor the growth, and refresh IEM every 2-3 days by aspirating the old medium and adding 500 µL of fresh IEM.
- Passage the organoids once they have recovered properly from thawing and have reached the right size to be processed (Figure 1C), as described in section 2.2.
- Passaging of intestinal organoids
NOTE: Chill the ECM on ice for at least 30 min, and keep the IEM at RT for at least 1 h before use.- Use the medium from one culture well to break up the organoid domes using a 1250 µL low-retention filter-tip, and transfer the well contents to a labeled 15 mL conical tube. Wash the well with 1 mL of BM, and transfer it to the same 15 mL conical tube.
- Repeat steps 2.2.1 and 2.2.2 with all other wells (a maximum of half a plate or 600 µL of ECM drops can be washed and added to one 15 mL conical tube).
- Add BM to fill the tube up to 12 mL, and pipet up and down 10x using a 10 mL pipette. Centrifuge at 85 × g for 5 min at 8 °C.
- Before removing the BM, check under the microscope to see whether all organoids are pelleted at the bottom of the 15 mL conical tube (Figure 1D). If there is no ECM overlaying the organoid pellet or the ECM layer is either clean or contains just debris, single cells, or very few organoids compared with the pelleted organoids, aspirate the supernatant and pipet out the ECM overlaying the organoid pellet very carefully using a P200 pipette.
NOTE: Organoids can get trapped in the ECM and do not sediment as a compact pellet due to low centrifugation force. If the ECM contains organoids, centrifuge the tube again at 450 × g for 5 min at 8 °C, and carefully remove the supernatant as described in 2.2.4. If there are multiple 15 mL conical tubes, they can be pooled after step 2.2.4. - Add 1 mL of BM to each pellet (of volume of 50-200 µL, depending on the organoid culture and density), and resuspend carefully. Pipet the organoids up and down at least 5x to shear them, avoiding foam formation. Check under the microscope to see whether the organoids are disrupted (Figure 2A). If the organoids are disrupted, proceed to step 2.2.7; if the organoids are not disrupted, pipet them another 5x. This time, touch the wall of the plastic tube with the pipette tip to exert more mechanical force to disrupt the organoids.
NOTE: Mechanical shearing of cystic (Figure 1C) and budding (Figure 1E) organoids is possible with either a 200 µL or 10 µL plastic pipette tip fitted on a low-retention 1250 µL filter-tip (Figure 1F), depending on the volume required for disrupting the organoids. The use of a narrowed glass pipette (Figure 1F) is recommended when more than 200 µL of ECM containing the organoids are processed (one well of a 6-well plate or 4 wells of a 24-well plate). - Check under the microscope again to see whether the organoids are disrupted. If disrupted, proceed with the next step; if not, pipet the organoids up to 20x, checking the organoids under the microscope regularly. If the organoids are still not disrupted, add 25% v/v cell dissociation reagent 1 (see the Table of Materials) to the suspension, incubate in the water bath at 37 °C for 2 min, and pipet the organoids up to 20x, checking the organoids under the microscope regularly to make sure they are not digested to single cells.
- Add up to 12 mL of BM to the 15 mL conical tube, and wash the organoid pellet by pipetting up and down. Centrifuge at 85 × g for 5 min at 8 °C. Discard the supernatant, and adjust the final concentration to 70% v/v ECM by adding IEM and ECM to the organoid pellet.
- Start resuspending the organoid pellet with double the volume of IEM/ECM collected for passaging, and seed 5 µL of the suspension to check the density. Continue plating if the density is appropriate (Figure 2B); add more IEM/ECM solution if the density is too high. Add 200 µL of the suspension to each well of a prewarmed 6-well plate, making separate drops of 10 µL volume.
- Turn the plate upside down, and leave it in the biosafety cabinet for 5 min. Transfer the plate still upside down to the 37 °C incubator, leaving it for another 30 min. Add 2 mL of IEM with 10 µM ROCK inhibitor to each well, and transfer the plate to the incubator.
- Image one drop regularly to monitor the growth, and refresh IEM every 2-3 days by aspirating the old medium and adding 2 mL of fresh IEM.
- Passaging of intestinal organoids for epithelial monolayer preparation
- Passage organoids 3 days prior to harvesting for monolayer preparation by following the same passaging protocol described in section 2.2 with one exception. In step 2.2.7, resuspend the organoids in 1-1.5x the starting volume of IEM/ECM to have a higher density and expansion potential when they are harvested for monolayer preparation (Figure 3A).
3. Epithelial monolayer preparation
- Culture epithelial monolayers on both 24-well and 96-well membrane inserts with a variety of available plate types (Table 1). Use high-throughput system (HTS) membrane inserts for both sizes as these contain an integral tray with the membrane inserts and a receiver plate. For the 24-well format, the use of plates with separate removable membrane inserts is also possible.
NOTE: Different membrane types (polyethylene terephthalate (PET) or polycarbonate) and pore sizes (0.4-8.0 µm) are available and can be used depending on experimental needs. Monolayers can only be imaged by brightfield when inserts with PET membranes are used. Light-tight membranes block fluorescent light leakage from the apical to the basolateral compartment and can be considered when dynamic transport or permeability of fluorescently labeled substrates is studied. The current protocol uses 24-well membrane inserts; adaptations for 96-well membrane inserts are described in section 5. Depending on the density, morphology, and size of the organoids (Figure 3A) , 6 wells of a 6-well plate (as seeded in section 2.3) are enough for seeding a full 24-well plate of membrane inserts. - Coating membrane inserts with ECM
NOTE: If there are doubts about having enough cells, coat the inserts after counting the cells. This is to prevent unnecessary coating and loss of the expensive membrane inserts.- Place the membrane inserts into the support plate in the biosafety cabinet. Dilute the ECM 40x in ice-cold Dulbecco's phosphate-buffered saline (DPBS) with Ca2+ and Mg2+, and pipet 150 µL of the diluted ECM into the apical compartment of each insert. Incubate the plate at 37 °C for at least 1 h.
- Preparation of cells for seeding
- Prewarm aliquots of the cell dissociation reagent 2 in the water bath (37 °C). Prepare 2 mL of the reagent for each well of a 6-well plate.
- Transfer the culture plate containing the organoids (prepared in section 2.3) from the incubator to the biosafety cabinet. Process the organoids, as described in steps 2.2.1.-2.2.4. Do not pool multiple tubes into one tube.
- Fill the tube, containing organoids from a maximum of 3 wells of a 6-well plate, up to 12 mL with DPBS (without Ca2+ and Mg2+), and pipet up and down 10x using a 10 mL pipette. Centrifuge at 85 × g for 5 min at 8 °C, and aspirate the supernatant without disturbing the organoid pellet.
- Add 2 mL of the prewarmed cell dissociation reagent 2 per well of a 6-well plate used as the starting material and resuspend. Incubate the tubes diagonally or horizontally for 5 min in the water bath at 37 °C, to prevent the sinking of the organoids to the bottom of the tube.
- Pipet up and down 10x using a 5 mL sterile plastic pipette or a P1000 pipette, depending on the total volume of the cell dissociation reagent. Check the organoid suspension under the microscope to see if a mixture of single cells and some cell clumps consisting of 2-4 cells has formed (Figure 3B). If needed, continue the digestion by repeating steps 3.3.4-3.3.5 (do not increase volume of cell dissociation reagent) until the mixture looks similar to Figure 3B.
NOTE: Avoid digesting the organoids fully to single cells. It is necessary to have some small groups of cells (i.e., groups of 2-4 cells). - Stop cell dissociation by adding up to 12 mL of BM including 10 µM ROCK inhibitor to the cell suspension. Centrifuge at 450 × g for 5 min at 8 °C, and aspirate the supernatant without disturbing the cell pellet. When handling the same organoid culture in several 15 mL conical tubes, pool the cell pellets and resuspend them in 12 mL of BM.
- Filter the cell suspension through a 40 µm strainer prewetted with BM, and harvest the flow-through into a 50 mL conical tube. Wash the strainer with 10 mL of BM, and harvest the flow-through into the same 50 mL conical tube.
- Transfer the strained cell suspension into two new 15 mL conical tubes. Centrifuge at 450 × g for 5 min at 8 °C, and aspirate the supernatant without disturbing the cell pellet. Resuspend the cells in 4 mL of IEM supplemented with 10 µM ROCK inhibitor per full culture plate used as starting material.
- Mix a small amount of cell suspension in a 1:1 ratio with trypan blue for counting. Count the live, not blue, cells (Figure 3C), and calculate the total number of live cells. In small clumps, count each individual cell.
- Prepare a cell suspension containing 3 × 106 live cells per mL of IEM supplemented with 10 µM ROCK inhibitor.
- Seeding cells on polyester membrane inserts
- Carefully aspirate DPBS from the ECM-coated inserts (step 3.2.1), whilst keeping the plate horizontally. Pipet 800 µL of IEM supplemented with ROCK inhibitor into each basolateral compartment. Pipet 150 µL of the cell suspension prepared in step 3.3.10 onto the ECM-coated membrane in the apical compartment dropwise. Per plate, be sure to have at least one "blank" well with BM only.
- Once the cells have sedimented onto the membrane, measure transepithelial electrical resistance (TEER), as described in section 4.1, and image the membrane inserts using a microscope. Place the plate in the incubator at 37 °C and 5% CO2. Measure TEER every day, and acquire images regularly to monitor monolayer formation (Figure 4A-D).
- Refreshing monolayers
NOTE: Refresh the medium every 2-3 days, adhering to the following order to maintain a positive hydrostatic pressure above the cells and prevent cells from being pushed off the membrane. While refreshing the medium, make sure the monolayer, which is visible upon aspiration of the medium, is not damaged by the pipette tip.- Remove the medium from the basolateral compartments of the plate containing the membrane inserts. Then, carefully aspirate the medium from the apical compartments of the membrane inserts.
- Add 150 µL of fresh IEM dropwise to each apical compartment, and then add 800 µL of fresh IEM to each basolateral compartment.
- Enrichment of the monolayer for desired intestinal epithelial cell types
- Allow the monolayer to become confluent in IEM, corresponding to a TEER value of around 100 Ω·cm² (as calculated in step 4.1.1.4). Check under the microscope to determine whether the monolayers have completely formed (Figure 4D) and for the absence of holes (as seen in Figure 4B,C).
- Carefully remove IEM from the basolateral and apical compartments of the membrane inserts, and replace with either eDM or cDM as prepared in section 1.4. Culture the monolayer for another 3-4 days in the specific differentiation medium to get the organoid cells enriched with the desired specific cell type. Refresh the medium every 2-3 days, as described in section 3.4.
- Measure TEER daily, and acquire images regularly if desired (Figure 5A-C).
NOTE: The TEER value that indicates a fully organized enriched monolayer varies per organoid culture; typically TEER values increase to 600 and can increase up to 1000 Ω·cm2 (as calculated in step 4.1.1.4) after 3 days in differentiation media and are stable for 3-5 days.
4. Epithelial monolayer assay readouts
- Measurement of transepithelial electrical resistance (TEER)
NOTE: TEER measurements are widely accepted as a method to analyze tight junction dynamics and barrier function integrity in biological models of physiological barriers, such as epithelial monolayers28, 29. Increase in TEER after differentiation because of increased cellular interaction at tight junctions can be measured using a manual TEER meter or an automated TEER measurement robot.- Measurement of TEER using a manual TEER meter
- Clean the electrode with 70% ethanol, and let it air-dry inside the biosafety cabinet. Place the electrode in a tube containing BM. Connect the electrode to the manual TEER meter. Turn the Function switch to measure in Ohms (Ω). Turn the Power switch on.
- Place the short electrode in the apical compartment of the insert, while the long electrode is positioned in the basolateral compartment (Figure 6A). Avoid touching the monolayer.
- Measure resistance in the blank well (Rblank), and then measure the remaining samples (Rsample) in the same way. Wash the electrode with BM between samples with different conditions. Clean the electrode first with demi water and then with 70% ethanol and let it air-dry.
- Calculate TEER (Ω·cm2): [Rsample (Ω) - Rblank (Ω)] × membrane area (cm2) (Table 1 and Figure 6B).
- Measurement of TEER using an automated TEER measurement robot (Table of Materials)
- Perform automated TEER measurements when using HTS systems for 96-well and 24-well HTS plates containing membrane inserts. Use different electrodes for TEER measurement for both types (24- and 96- HTS membrane inserts). To measure TEER using an automated TEER measurement robot, follow the manufacturer's instructions.
- Measurement of TEER using a manual TEER meter
- Measurement of epithelial barrier integrity and permeability
NOTE: This protocol introduces Lucifer Yellow permeability from the apical to basolateral compartment as an indication of monolayer integrity. This section describes fluorescence measurement in the basolateral compartment after a 1 h incubation step to evaluate monolayer permeability and thus, barrier integrity. This measurement is an end-point assay and is especially useful when testing compounds for their effect on barrier integrity.- Thaw Lucifer Yellow on ice, and let BM equilibrate to RT. For one 24-well plate of membrane inserts, prepare 5 mL of working solution of 60 µM Lucifer Yellow in BM.
NOTE: Lucifer Yellow is light-sensitive. Prepare dilutions in dark 1.5 mL sterile tubes and perform all steps with the biosafety cabinet light switched off. - Carefully remove the medium from the basolateral and apical compartments of the membrane inserts, as described in step 3.5.1. If desired, scratch one untreated monolayer using a pipette tip as a positive control for Lucifer Yellow leakage through a damaged barrier.
- Add 150 µL of BM with 60 µM Lucifer Yellow to each apical compartment, and add 800 µL of BM without Lucifer Yellow to each basolateral compartment. Place the plate on a shaker at 37 °C, 50 rpm for 60 min.
- In the meantime, prepare a standard curve of Lucifer Yellow in BM starting with the working solution prepared in step 4.2.1. Dilute 1:3 in each step until a concentration of 3 nM is reached. Include a negative control (BM only).
- Transfer 100 µL of each standard in triplicate to a 96-well transparent plate. After 60 min incubation, remove the membrane inserts and transfer 100 µL from each basolateral well (step 4.2.3) in triplicate to the 96-well transparent plate. Measure fluorescence of the plate using a plate reader at an excitation wavelength of 430 nm and an emission wavelength of 530 nm.
- After correcting for the negative control value (BM only), use the standard curve values to calculate the Lucifer Yellow concentration in the basolateral compartment (final receiver concentration (µM)).
- Calculate the apparent permeability coefficient (Papp) according to the following formula (Figure 6C):

- For a 24-well plate containing membrane inserts, use the following formula:
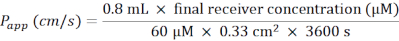
- Thaw Lucifer Yellow on ice, and let BM equilibrate to RT. For one 24-well plate of membrane inserts, prepare 5 mL of working solution of 60 µM Lucifer Yellow in BM.
- Fixing monolayers and preparing paraffin blocks for histology
NOTE: Epithelial monolayers can be used for histologic staining for evaluation of their cellular composition, polarity, and expression of different proteins of interest such as junctional proteins, proliferation, or differentiation markers. This section describes paraffin block preparation for histologic staining.- Carefully remove the medium from the basolateral and apical compartments of the membrane inserts, as described in steps 3.5.1.
- Wash the monolayers by adding 150 µL of DPBS (without Ca2+ and Mg2+) to each apical compartment and 800 µL to each basolateral compartment. Carefully aspirate the DPBS again, first from the basolateral compartment and then from the apical compartment.
NOTE: The basolateral compartment will stay empty from this step on. - In a fume hood, add 150 µL of 4% paraformaldehyde to each apical compartment, and incubate for 30 min at RT.
NOTE: From this step onwards, perform all actions in this section inside a fume hood, as paraformaldehyde is toxic. - Carefully aspirate the fixative from the apical compartments of the membrane inserts, and dispose of it as liquid halogen waste.
NOTE: From this step onwards, dispose of all liquid waste as liquid halogen waste. - Wash the monolayers by adding 200 µL of DPBS (without Ca2+ and Mg2+) to each apical compartment, and carefully aspirate the DPBS again. Repeat this step one more time.
- Add 200 µL of 25% ethyl alcohol (EtOH) to each apical compartment, and incubate for 15 min at RT. After 15 min, carefully aspirate the 25% EtOH from the apical compartments of the membrane inserts. Repeat with 50% EtOH solution and subsequently with 70% EtOH solution.
- Add 200 µL of 70% EtOH to each apical compartment, and wrap the plate with parafilm. Store it at 4 °C until further use.
- Carefully aspirate the 70% EtOH, and use a scalpel to carefully cut the monolayer membranes from the inserts. Cut from the basolateral side, around the edge of the insert.
- Prepare paraffin blocks following the standard procedure.
- When the paraffin is still warm, take the monolayer from the paraffin with tweezers, and place it on a precooled surface.
- Be careful not to damage the monolayer. Cut the monolayer in half using a single edge blade.
- When the paraffin in the bottom of the cassette starts to solidify, use heated tweezers to place the two monolayer parts in the paraffin, next to each other with the straight side down and in a vertical direction to ensure that the monolayer will be vertical in the coupe.
- When paraffin blocks are ready, cut the blocks using a microtome, and make slides of 4 µm thick sections following standard procedure. Make sure that the monolayers end up vertical in the coupe.
- Perform histologic stains as described previously7,9. Use hematoxylin and eosin (H&E), Ki67, mucin-2 (MUC2), and Alcian Blue to show general morphology, proliferative cells, mucus production and goblet cells, respectively (Figure 6E).
NOTE: Additional differentiation markers, such as lysozyme for Paneth cells, can be used as well. This marker is not presented in Figure 6E as Paneth cells are present in small intestinal epithelium rather than colon epithelium.
- Secreted protein measurement in medium supernatant
- Measure lysozyme levels in the apical supernatant of ileal monolayers (see Figure 6D) using the kit listed in the Table of Materials. If desired, measure levels of different cytokines and other proteins of interest.
- Gene expression analysis
- Quantify the effects of the differentiation media on the expression of epithelial cell marker genes using quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
- Lyse the monolayers in 350 µL of RNA lysis buffer followed by RNA isolation according to the manufacturer's instructions. Perform cDNA synthesis and qPCRs, as described earlier7,9, using the cDNA synthesis kits, master mix, and oligonucleotides listed in the Table of Materials.
- Quantify the effects of the differentiation media on the expression of epithelial cell marker genes using quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
5. Upscaling to 96-well plates containing membrane inserts
NOTE: Prepare epithelial monolayers for higher throughput drug screenings or multiple medium conditions using HTS 96-well plates containing membrane inserts.
- Adaptations when preparing monolayers in 96-well format
- Follow all steps described in this protocol for 24-well plates containing membrane inserts, changing volumes and cell numbers to those described in Table 1. For preparing monolayers on 96-well plates with membrane inserts, proceed as described in section 3 with the following differences.
- Approximately 9 wells of a 6-well culture plate with organoid density represented in Figure 3A are needed to seed a full 96-well plate with membrane inserts. In step 3.2.1, precoat the membranes with 67 µL of 40x diluted ECM in DPBS (with Ca2+ and Mg2+).
- In section 3.5, first transfer the integral plate of membrane inserts to another 96-well plate to allow medium refreshment of both apical and basolateral compartments.
- Follow all steps described in this protocol for 24-well plates containing membrane inserts, changing volumes and cell numbers to those described in Table 1. For preparing monolayers on 96-well plates with membrane inserts, proceed as described in section 3 with the following differences.
Results
Figure 1A shows a representative brightfield image of intestinal organoids after thawing them from a cryovial. It is important to thaw organoids at a high density to ensure optimal recovery. Organoids are plated in 24- or 6-well plates in ECM domes of approximately 10 µL (Figure 1B). Most normal intestinal organoids have a cystic morphology. After recovering from the thawing process, the organoids grow to a bigger size and are ready to be passaged after 3-7...
Discussion
This protocol describes the general manipulation and maintenance of intestinal organoids as well as the preparation and possible applications of epithelial monolayers derived from these organoids. To date, monolayers have been successfully prepared from the duodenum, ileum, and different regions of colon organoids derived from normal as well as previously and actively inflamed intestinal tissue (unpublished data). The application of patient-derived organoid monolayers facilitates the study of barrier function in a diseas...
Disclosures
The authors declare no conflict of interest.
Acknowledgements
This work is supported by the Topsector Life Sciences & Health - Topconsortium voor Kennis en Innovatie Health~Holland (LSH-TKI) public-private partnerships (PPP) allowance of the Dutch LSH sector with Project number LSHM16021 Organoids as novel tool for toxicology modelling to Hubrecht Organoid Technology (HUB) and HUB internal funding to Disease Modeling and Toxicology department. We thank the laboratories of Sabine Middendorp (Division of Pediatric Gastroenterology, Wilhelmina Children's Hospital, UMC, Utrecht) and Hugo R. de Jonge and Marcel J.C. Bijvelds (Department of Gastroenterology and Hepatology, Erasmus MC, Rotterdam) for providing initial technical support to set up monolayers on membrane inserts.
Materials
| Name | Company | Catalog Number | Comments |
| 100% ethanol | Fisher Emergo | 10644795 | |
| 1250, 300, and 20 µL low-retention filter-tips | Greiner bio-one | 732-1432 / 732-1434 / 732-2383 | |
| 15 mL conical tubes | Greiner bio-one | 188271 | |
| 24-well cell culture plates | Greiner bio-one | 662160 | |
| 24-well HTS Fluoroblok Transwell plate (light-tight) | Corning | 351156 | |
| 24-well HTS Transwell plates (Table 1) | Corning | 3378 | |
| 24-well plate with Transwell inserts | Corning | 3470 | |
| 40 µm cell strainer | PluriSelect | 43-50040-01 | |
| 50 mL conical tubes | Greiner bio-one | 227261 | |
| 6-well cell culture plates | Greiner bio-one | 657160 | |
| 96-well black plate transparent bottom | Greiner bio-one | 655090 | |
| 96-well fast thermal cycling plates | Life Technologies Europe BV | 4346907 | |
| 96-well HTS Fluoroblok Transwell plate | Corning | 351162 | |
| 96-well HTS Transwell plates (Table 1) | Corning | 7369 | |
| 96-well transparent culture plate | Greiner bio-one | 655180 | |
| A83-01 | Bio-Techne Ltd | 2939 | |
| Accutase Cell Dissociation Reagent | Life Technologies Europe BV | A11105-01 | Cell dissociation reagent 2 |
| Advanced DMEM/F-12 | Life Technologies Europe BV | 12634028 | |
| B27 supplement | Life Technologies Europe BV | 17504001 | |
| Cell culture microscope (light / optical microscope) | Leica | ||
| CellTiter-Glo | Promega | G9683 | |
| Centrifuge | Eppendorf | ||
| CO2 incubator | PHCBI | ||
| DAPT | Sigma-Aldrich | D5942 | |
| DEPC treated H2O | Life Technologies Europe BV | 750024 | |
| Dulbecco's phosphate-buffered saline (DPBS) with Ca2+ and Mg2+ | Life Technologies Europe BV | 14040091 | |
| DPBS, powder, no calcium, no magnesium | Life Technologies Europe BV | 21600069 | |
| EnzChek Lysozyme Assay Kit | Life Technologies Europe BV | E22013 | |
| EVOM2 meter with STX electrode | WTI | ||
| Gastrin | Bio-Techne Ltd | 3006 | |
| Glass pipettes | Volac | ||
| GlutaMAX | Life Technologies Europe BV | 35050038 | |
| hEGF | Peprotech | AF-100-15 | |
| HEPES | Life Technologies Europe BV | 15630056 | |
| Human Noggin | Peprotech | 120-10C | |
| Human Rspo3 | Bio-Techne Ltd | 3500-RS/CF | |
| IWP-2 | Miltenyi Biotec | 130-105-335 | |
| Ki67 primary antibody | Sanbio | BSH-7302-100 | |
| Ki67 secondary antibody | Agilent | K400111-2 | |
| Kova International Glasstic Slide with Counting grids | Fisher Emergo | 10298483 | |
| Laminar flow hood | Thermo scientific | ||
| Lucifer Yellow CH dilithium salt | Sigma-Aldrich | L0259 | |
| Matrigel, Growth Factor Reduced (GFR) | Corning | 356231 | extracellular matrix (ECM) |
| MicroAmp Fast 8-Tube Strip, 0.1 mL | Life Technologies Europe BV | 4358293 | |
| MicroAmp Optical 8-Cap Strips | Life Technologies Europe BV | 4323032 | |
| Microcentrifuge tubes | Eppendorf | 0030 120 086 | |
| Micropipettes (1000, 200, and 20 µL) | Gilson | ||
| Microtome | Leica | ||
| MUC2 primary antibody | Santa Cruz Biotechnology | sc-15334 | |
| MUC2 secondary antibody | VWR | VWRKS/DPVR-HRP | |
| Multichannel pipette (200 µL) | Gilson | ||
| N-acetylcysteine | Sigma-Aldrich | A9165 | |
| NGS Wnt | U-Protein Express | N001-0.5mg | |
| Nicotinamide | Sigma-Aldrich | N0636 | |
| Oligonucleotide ALPI1/Forward | Custom-made | GGAGTTATCCTGCTCCCCAC | |
| Oligonucleotide ALPI1/Reverse | Custom-made | CTAGGAGGTGAAGGTCCAACG | |
| Oligonucleotide LGR5/Forward | Custom-made | ACACGTACCCACAGAAGCTC | |
| Oligonucleotide LGR5/Reverse | Custom-made | GGAATGCAGGCCACTGAAAC | |
| Oligonucleotide MUC2/Forward | Custom-made | AGGATCTGAAGAAGTGTGTCACTG | |
| Oligonucleotide MUC2/Reverse | Custom-made | TAATGGAACAGATGTTGAAGTGCT | |
| Oligonucleotide TBP/Forward | Custom-made | ACGCCGAATATAATCCCAAGCG | |
| Oligonucleotide TBP/Reverse | Custom-made | AAATCAGTGCCGTGGTTCGTG | |
| Optical adhesive covers | Life Technologies Europe BV | 4311971 | |
| PD0325901 | Stemcell Technologies | 72184 | |
| Penicillin/streptomycin | Life Technologies Europe BV | 15140122 | |
| Plate shaker | Panasonic | ||
| PowerUp SYBR Green Master Mix | Fisher Emergo | A25776 | |
| Primocin | InvivoGen | ANT-PM-2 | antimicrobial formulation for primary cells |
| Qubit RNA HS Assay Kit | Life Technologies Europe BV | Q32852 | |
| Reagent reservoir for multichannel pipet | Sigma-Aldrich | CLS4870 | |
| REMS AutoSampler with 24-probe or 96C-probe | WTI | ||
| Richard-Allan Scientific Alcian Blue/PAS Special Stain Kit | Thermo scientific | 87023 | |
| RNase-Free DNase Set | Qiagen | 79254 | |
| RNeasy Mini Kit | Qiagen | 74106 | |
| SB202190 | Sigma-Aldrich | S7076 | |
| Serological pipettes | Greiner bio-one | 606180 / 607180 / 760180 | |
| Serological pipettor (Pipet-Aid) | Drummond | ||
| Single edge razor blade | GEM Scientific | ||
| Superscript 1st strand system for RT-PCR | Life Technologies Europe BV | 11904018 | |
| Tecan Spark 10M plate reader | Tecan | ||
| Trypan Blue Solution, 0.4% | Life Technologies Europe BV | 15250-061 | |
| TrypLE Express Enzyme (1x) | Life Technologies Europe BV | 12605-010 | Cell dissociation reagent 1 |
| Water bath | Grant | ||
| Y27632 (ROCK inhibitor) | AbMole | M1817 |
References
- Haegebarth, A., Clevers, H. Wnt signaling, lgr5, and stem cells in the intestine and skin. The American Journal of Pathology. 174 (3), 715-721 (2009).
- Schoultz, I., Keita, &. #. 1. 9. 7. ;. V. The intestinal barrier and current techniques for the assessment of gut permeability. Cells. 9 (8), 1909 (2020).
- Martínez-Maqueda, D., Miralles, B., Recio, I., Verhoeckx, K. HT29 Cell Line. The Impact of Food Bio-Actives on Gut Health: In Vitro and Ex Vivo Models. , 113-124 (2015).
- Sato, T., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459 (7244), 262-265 (2009).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 141 (5), 1762-1772 (2011).
- Huch, M., et al. In vitro expansion of single Lgr5(+) liver stem cells induced by Wnt-driven regeneration. Nature. 11 (2), 179-194 (2013).
- Sachs, N., et al. Long-term expanding human airway organoids for disease modeling. The EMBO Journal. 38 (4), 1-20 (2019).
- Karthaus, W. R., et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 159 (1), 163-175 (2014).
- Boj, S. F., et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 160 (1-2), 324-338 (2015).
- Sachs, N., et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 172 (1-2), 373-386 (2018).
- Vlachogiannis, G., et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 359 (6378), 920-926 (2018).
- Van De Wetering, M., et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 161 (4), 933-945 (2015).
- Driehuis, E., et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proceedings of the National Academy of Sciences of the United States of America. 116 (52), 26580-26590 (2019).
- Tiriac, H., et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discovery. 8 (9), 1112-1129 (2018).
- d'Aldebert, E., et al. Characterization of human colon organoids from inflammatory bowel disease patients. Frontiers in Cell and Developmental Biology. 8, 363 (2020).
- Dotti, I., et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 66 (12), 2069-2079 (2017).
- VanDussen, K. L., et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 64 (6), 911-920 (2015).
- Noel, G., et al. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Scientific Reports. 7, 45270 (2017).
- Bartfeld, S., et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 148 (1), 126-136 (2015).
- van Es, J. H., et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nature Cell Biology. 7 (4), 381-386 (2005).
- van Es, J. H., et al. Dll1 marks early secretory progenitors in gut crypts that can revert to stem cells upon tissue damage. Nature Cell Biology. 14 (10), 1099-1104 (2012).
- de Lau, W. B. M., Snel, B., Clevers, H. C. The R-spondin protein family. Genome Biology. 13 (3), 1-10 (2012).
- Basak, O., Beumer, J., Wiebrands, K., Seno, H., van Oudenaarden, A., Clevers, H. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell. 20 (2), 177-190 (2017).
- Beumer, J., et al. Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nature Cell Biology. 20 (8), 909-916 (2018).
- Yin, X., Farin, H. F., van Es, J. H., Clevers, H., Langer, R., Karp, J. M. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nature Methods. 11 (1), 106-112 (2014).
- Boj, S. F., et al. Forskolin-induced swelling in intestinal organoids: An in vitro assay for assessing drug response in cystic fibrosis patients. Journal of Visualized Experiments. (120), (2017).
- Miao, Y., et al. Next-generation surrogate Wnts support organoid growth and deconvolute Frizzled pleiotropy in vivo. Cell Stem Cell. 27 (5), 840-851 (2020).
- Srinivasan, B., et al. TEER measurement techniques for in vitro barrier model systems. Journal of Laboratory Automation. 20 (2), 107-126 (2015).
- Blume, L. -. F., Denker, M., Gieseler, F., Kunze, T. Temperature corrected transepithelial electrical resistance (TEER) measurement to quantify rapid changes in paracellular permeability. Die Pharmazie. 65 (1), 19-24 (2010).
- Lea, T., Verhoeckx, K., et al. Caco-2 cell line. The Impact of Food Bio-Actives on Gut Health: In Vitro and Ex Vivo Models. , 103-111 (2015).
- Heo, I., et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nature Microbiology. 3 (7), 814-823 (2018).
- Watanabe, K., et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology. 25 (6), 681-686 (2007).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved