Method Article
Monitoring Protein-Ligand Interactions in Human Cells by Real-Time Quantitative In-Cell NMR using a High Cell Density Bioreactor
In This Article
Summary
This protocol describes the setup of an NMR bioreactor to keep encapsulated human cells viable for up to 72 h, followed by time-resolved in-cell NMR data acquisition and analysis. The methodology is applied to monitor intracellular protein-ligand interactions in real time.
Abstract
In-cell NMR is a unique approach to observe the structural and dynamic properties of biological macromolecules at atomic resolution directly in living cells. Protein folding, chemical modifications, and conformational changes induced by ligand binding can be observed. Therefore, this method has great potential in the context of drug development. However, the short lifetime of human cells confined in the NMR spectrometer limits the application range of in-cell NMR. To overcome this issue, NMR bioreactors are employed that can greatly improve the cell sample stability over time and, importantly, enable the real-time recording of in-cell NMR spectra. In this way, the evolution of processes such as ligand penetration and binding to the intracellular protein target can be monitored in real time. Bioreactors are often limited by low cell viability at high cell numbers, which results in a trade-off between the overall sensitivity of the experiment and cell viability. We recently reported an NMR bioreactor that maintains a high number of human cells metabolically active for extended periods of time, up to 72 h. This setup was applied to monitor protein-ligand interactions and protein chemical modification. We also introduced a workflow for quantitative analysis of the real-time NMR data, based on multivariate curve resolution. The method provides concentration profiles of the chemical species present in the cells as a function of time, which can be further analyzed to obtain relevant kinetic parameters. Here we provide a detailed description of the NMR bioreactor setup and its application to monitoring protein-ligand interactions in human cells.
Introduction
In-cell Nuclear Magnetic Resonance (NMR) spectroscopy has recently emerged as a powerful approach to investigate structural and dynamical properties of macromolecules within the cellular environment1,2,3,4,5,6. In-cell NMR succeeded in the investigation of functionally relevant processes such as protein folding/misfolding7,8,9, metal binding7,10, disulfide bond formation11,12, and protein−protein interaction13, protein-ligand interaction14,15,16, and nucleic acid-ligand interaction17,18 in living human cells. One of the limiting factors of in-cell NMR applications is the short lifetime of the cells during the experiment. The solution to this problem involves the use of NMR bioreactors. In these devices, a constant flow of growth medium is applied to the cells, which are kept confined within the NMR spectrometer, in order to provide oxygen and nutrients and to remove toxic byproducts. Following the advent of in-cell NMR, several NMR bioreactors designs have been developed to improve cell viability for longer periods of time, in which either bacteria or mammalian cells are encapsulated in a hydrogel19,20,21,22 or kept in suspension and perfused through the use of a microdialysis membrane23. Such bioreactors have allowed the acquisition of longer NMR experiments with increased signal-to-noise ratio (S/N)5 and, even more importantly, could be employed to investigate cellular processes in real time22,23,24. Thanks to the high chemical and conformational sensitivity of NMR, the latter application can provide precious insights on the kinetics of functional processes within living cells at atomic resolution.
In this protocol, we show how to set up and operate an improved bioreactor recently reported25, which was obtained by combining an existing modular bioreactor design23 with the approach relying on cell encapsulation in hydrogel that were pioneered by other groups19,20,21,22,26,27. We describe the application of the bioreactor to real-time in-cell NMR studies of intracellular protein-observe ligand binding in HEK293T cells. In the bioreactor, cells are encapsulated at high density in agarose gel threads and are maintained highly viable and metabolically active for up to 72 h, during which real-time in-cell NMR experiments are recorded. The bioreactor is composed of a glass tube that fits standard 5 mm NMR probes that is watertight and connected to a tube holder so that the internal sample chamber has 4.2 mm internal diameter, 38 mm height, and a volume of 526 µL. The inlet is a 7-meter-long PEEK capillary (o.d. = 1/32", i.d. = 0.5 mm) inserted in the sample chamber down to ~6 mm from the bottom, while the outlet is a 7-meter-long PTFE capillary (o.d. = 1/32", i.d. = 0.5 mm) attached at the top of the tube holder (Figure 1). The tubing is coaxially inserted in a temperature-controlled line connected to a water bath. The inlet and outlet are connected through PEEK tubing to a 4-way, 2-position valve attached to an FPLC pump for controlling the medium flow and a waste container.
The bioreactor is applied to study the kinetics of the interaction, previously reported14,25, between two drugs, acetazolamide (AAZ) and methazolamide (MZA), in human cells with the second isoform of human carbonic anhydrase (CA II), a pharmacologically relevant target28,29,30, and the kinetics of the formation of the intramolecular disulfide bond, promoted by the small molecule ebselen25,31, of the copper-free, zinc-bound form of human copper, zinc superoxide dismutase (SOD1), an antioxidant enzyme implicated in the onset of amyotrophic lateral sclerosis7,8,32. Finally, quantitative analysis of the real-time NMR data is performed in MATLAB using the Multivariate Curve Resolution-Alternating Least Squares (MCR-ALS) algorithm33, through which the pure spectral components and the concentration profiles as a function of time are obtained for the species observed, that can be further analyzed to obtain relevant kinetic parameters.
The protocol starts from a T75 flask of HEK293T cells (~3 x 107 cells per flask) transiently overexpressing either human CA II (unlabeled) or human SOD1 (15N-labeled). The cells were grown and maintained in T75 flasks with DMEM high glucose by 1:10 passages every 3-4 days and transfected with the cDNA encoding the protein of interest 48 h prior to the experiment. The steps involved in this phase are reported in detail elsewhere34.
Protocol
1. Reagent and solution setup
- To prepare complete DMEM, add 5 mL L-glutamine 200 mM, 5 mL penicillin-streptomycin 100x, and 50 mL fetal bovine serum (FBS, 10% vol/vol final concentration) to 440 mL DMEM.
NOTE: This solution can be stored at 4 °C for 1 month. - Prepare agarose solution by dissolving 150 mg of low-gelling agarose in 10 mL of phosphate buffered saline (PBS) at 85 °C to obtain a 1.5% (w/v) solution. Sterilize by filtration with a 0.22 µm filter. Prepare 1 mL aliquots of agarose solution in 1.5 mL capped tubes and store at 4 °C.
- Prepare the bioreactor medium.
- Dissolve 13.4 g of DMEM powder in 1 L of ultrapure H2O.
NOTE: Depending on the application, the required final volume may differ (e.g., for 500 mL medium, dissolve 6.7 g of powder in 500 mL H2O). - Add 2% FBS, 10 mM NaHCO3, 1x penicillin-streptomycin (100x), and 2% D2O (e.g., for 500 mL medium, add 10 mL of FBS, 0.4 g of NaHCO3, 5 mL of penicillin-streptomycin 100x, and 10 mL of D2O).
- Measure the pH using a pH-meter and if needed adjust to 7.4 by adding HCl.
NOTE: Typically, the initial pH is very close to 7.4. - Filter the bioreactor medium with a vacuum-driven sterile filter in a sterile 250 mL or 500 mL glass bottle.
- In the laminar flow hood, seal the bottle with a sterile steel headpiece with two hose nozzles and connect them to a FEP tubing (o.d. = 1/8", i.d. = 1.6 mm) that will be connected to the pump and to a 0.22 µm PTFE syringe filter for air intake.
- Dissolve 13.4 g of DMEM powder in 1 L of ultrapure H2O.
2. Bioreactor setup
- Assemble the flow unit using a second flow unit NMR tube, which will be later replaced with the one containing the cells. Refer to the flow unit operating instructions for the correct assembly.
NOTE: At this point the flow unit should be already cleaned (if not, perform step 4.2). - Set the water bath connected to the flow unit temperature control to 37 °C. Place the reservoir bottle in the water bath.
- Connect the FEP tubing of the reservoir bottle to the pump.
- Turn the bioreactor valve to "bypass" and prefill the pump with medium.
- Turn the bioreactor valve to "flow" and prefill the bioreactor with medium at 0.1 mL/min.
3. Preparation of the cell sample
- Collect the cells from the CO2 incubator.
- Take a T75 flask of transfected HEK293T cells from the CO2 incubator and remove the spent medium.
- Wash the cells twice with 7 mL (each) of PBS at room temperature (~20 °C).
- Use 2 mL of trypsin/EDTA to detach cells. After adding the solution, incubate for 5 min at room temperature to detach the cells.
NOTE: Transfected cells may take slightly longer to get detached. If necessary, incubate the cells at 37 °C. - Inactivate trypsin with 20 mL of complete DMEM; thoroughly resuspend the cells by pipetting up and down and transfer them in a 50 mL centrifuge tube.
- Centrifuge the cells at 800 x g for 5 min at room temperature and discard the supernatant.
- Wash the cells with 10 mL of PBS at room temperature to remove the residual medium.
- Centrifuge the cells at 800 x g for 5 min at room temperature and discard the supernatant.
- Transfer the cell pellet to a 1.5 mL capped microcentrifuge tube.
- Embed cells in agarose threads.
- Melt one aliquot of solidified agarose at 85 °C in a water bath and subsequently keep it in solution at 37 °C in a block heater.
- With a Pasteur pipette, fill the bottom of the flow unit NMR tube with 60-70 µL of 1.5% agarose gel and place it in ice. This will create a ~5 mm high bottom plug that allows placing the cell sample within the active volume of the 1H NMR coil.
- Heat up the pellet of cells obtained in step 3.1.8 at 37 °C for 15−20 s in the thermoblock.
- Resuspend cells in 450 µL of agarose solution. Be careful to avoid the formation of bubbles.
- Aspirate the cell−agarose suspension into a ~30 cm long chromatography PEEK tubing (i.d. = 0.75 mm) connected to a 1 mL syringe.
NOTE: Before aspiration, the tubing and the dead volume of the syringe should be prefilled with PBS at room temperature to avoid the formation of bubbles. The length of the tubing is not critical. - Let the tubing cool down at room temperature for 2 min.
- Prefill the flow unit NMR tube with 100 µL of PBS at room temperature.
- Cast threads of cells embedded in agarose into the flow unit NMR tube by gently pushing the syringe.
NOTE: To fill the NMR tube homogenously, start by placing the end of the PEEK tubing at the bottom of the NMR tube and proceed towards the top while slowly swinging left-right. - Repeat steps 3.2.5, 3.2.6, and 3.2.8 until all the cell-agarose suspension has been cast.
- Insert cells in the bioreactor.
- Remove the empty NMR tube from the flow unit and increase the flow rate to 2 mL/min for a few minutes to remove residual gas bubbles in the inlet tubing.
- Set the flow rate to 0.2 mL/min and insert the NMR tube containing the cells by pushing it upwards slowly but steadily.
NOTE: The active flow of medium avoids the backflow of tube content through the inlet, that would otherwise occur during the insertion.
4. Bioreactor operation and cleaning
- Bioreactor operation during the NMR experiment.
- Set the temperature in the NMR spectrometer to 310 K.
- Insert the flow unit in the spectrometer.
- Supply the bioreactor medium at a flow rate of 0.1 mL/min for the whole duration of the in-cell NMR experiments.
- At the desired time during the experiment, inject a concentrated solution of external molecule to the medium reservoir bottle by piercing the silicone tubing with a sterile long-needle syringe.
NOTE: The final concentration of molecule in the medium should be chosen based on previous knowledge of cell toxicity and, if available, on the predicted/estimated diffusion rate through the cell membrane. - At the end of the NMR experiment, replace the tube containing the cells with an empty tube and rinse the flow unit with water.
- Bioreactor clean-in-place.
- Clean the flow unit by flowing the following solutions at 1 mL/min: 0.2 M sodium hydroxide (NaOH); 3 M citric acid; 0.2 M NaOH, for at least 30 min each, followed by sterile-filtered ultrapure water for >2 h.
- Clean and autoclave the reservoir bottle and tubing assembly after each run.
5. NMR experiments
- Setup of the NMR experiments.
NOTE: Perform these steps beforehand, prior to the preparation of the in-cell NMR sample, to avoid any delays between cell collection and data acquisition.- Create a new dataset at the NMR spectrometer and set the parameters for the desired NMR experiments.
- Set parameters for 1D 1H NMR experiments.
- Center the 1H carrier frequency at 4.7 ppm on the water signal.
- Select the zgesgp pulse program, set the spectral width to 20 ppm, and a 1,000-µs 180° square pulse for water suppression. Set an inter-scan delay of 1 s. Acquire the spectrum with 32 scans.
- For cells expressing unlabeled CA II: select the p3919gp pulse program, set the spectral width to 30 ppm to cover the imino region of the spectrum, and adjust the delay for binomial water suppression so that the maximum excitation is centered at the chemical shifts of the signals of interest (d7 = 20 µs at 950 MHz). Set an inter-scan delay of ≥1 s. Acquire with 512 scans.
- For cells expressing 15N-labeled SOD1: select the sfhmqf3gpph pulse program, set the 1H and 15N spectral widths to 16 and 50 ppm, respectively, the shaped pulse offset and excitation bandwidth to 8.5 and 6 ppm, respectively, and a 350 µs pulse for decoupling scheme (garp4 or other depending on the instrument). Set an inter-scan delay of 0.3 s. Acquire with 16 scans and 128 increments in the 15N dimension.
- Real-time NMR spectra acquisition.
- Once the bioreactor is inserted in the NMR spectrometer, wait for a few minutes to allow the exchange of the medium.
NOTE: This process is easily monitored from the appearance of the lock signal as the PBS is replaced with medium containing 2% D2O. - Adjust the matching and tuning of the 1H channel, shim the magnet, and calculate the 1H 90° hard pulse length.
- Adjust the 1H power levels in each pulse sequence according to the 1H hard pulse.
- Record a first zgesgp 1H spectrum to check the sample content and the field homogeneity.
- Copy the zgesgp and the p3919gp/sfhmqcf3gpph experiments to the desired number and queue them in the acquisition spooler.
NOTE: The zgesgp spectra are only used to control the state of the sample and the field homogeneity; therefore, they can be either skipped or recorded less frequently. - For cells expressing unlabeled CA II: process the p3919gp spectra by applying zero filling and exponential line broadening window function (LB = 20 Hz).
- For cells expressing 15N-labeled SOD1: process the sfhmqcf3gpph spectra by applying zero filling and squared sine bell window function (SSB = 2) in both dimensions.
NOTE: The size of the processed spectra can be further reduced by removing regions free of signals (in Topspin, this is done by setting the desired STSR and STSI values).
- Once the bioreactor is inserted in the NMR spectrometer, wait for a few minutes to allow the exchange of the medium.
6. MCR-ALS analysis
- For the analysis of CA II spectra, import 1D spectral regions in MATLAB R2019b.
- In the software, create a list of experiments to be exported in the Process Dataset List menu.
- Using a modified version of the au program convbin2asc, export the spectral region of interest in ASCII format for each spectrum.
NOTE: This creates a text file named ascii-spec.txt in each spectrum subdirectory. - In MATLAB, import the spectral regions using the custom script Load_ascii_spectra.
NOTE: This script requires the dataset directory as an input and produces a 2D array spectra containing the stacked 1D spectra and a 1D array cs containing the chemical shifts. - Run the Load_acqus script to extract the timestamps from the 1D spectra.
NOTE: This script produces a 1D array times_hours containing the time increment for each spectrum expressed in hours, with the initial spectrum at time = 0.
- For the analysis of SOD1 spectra, import 2D spectra in MATLAB R2020b.
- In Topspin, create a list of experiments to be exported in the Process Dataset List menu.
- In MATLAB, import the 2D spectra using the custom script Load_2D_spectra.
NOTE: This script requires the dataset directory as an input and produces a 3D array Spectra containing the stacked 2D spectra and a 1D array cs containing the chemical shifts. - Run the Load_acqus script to extract the timestamps from the 2D spectra.
- Specify the spectral regions of interest in the custom script Cut_2D_spectra and run the script to cut 3D sub-arrays [(1H x 15N) spectral intensities) x time]; reshape them as 2D arrays (time points x spectral intensities) and join them together.
NOTE: This produces a 2D array JoinSpec_flat containing the reshaped and joined spectral regions.
- Run MCR-ALS 2.0 in GUI mode.
- Open MCR-ALS 2.0 GUI by running the mcr_main script.
- In the Data Selection tab, load the spectra or the JoinSpec_flat matrix. The data can be plotted for checking.
- Evaluate the number of components either by Singular Value Decomposition (SVD) or manually.
NOTE: The number of components should correspond to the number of distinct species present in the experiment. In this case n = 2, corresponding to the free and bound protein. - Select a method for the initial estimation of the pure spectra. Either purest variable detection or Evolving Factor Analysis (EFA) can be used.
- In the Selection of the Data Set window, select Continue.
- Set the constraints for the concentrations in the Constraints: Row Mode window. Apply a non-negativity constraint, select fnnls (Fast nonnegativity-constrained least-squares) as "implementation and 2 species". Apply 1 closure constraint, set the constraint to 1, the closure condition as "equal to" and apply to all species.
NOTE: This forces the sum of each species concentrations to be equal to 1, so that the obtained profiles of each species are normalized with respect to the total protein concentration. - Set the constraints for the spectra in the Constraints: Column Mode window. Apply a non-negativity constraint, select fnnls as "implementation and 2 species".
NOTE: This constraint should not be applied if negative signals are present in the NMR spectra. - In the final window, set 50 iterations and 0.01 convergence criterion. Specify the output names for Concentrations, Spectra, and Std. deviation. Click on Continue to run the MCR-ALS fitting.
NOTE: A graphical window will show the result of the fitting with plots of the concentration profiles and the spectra of the pure components. - For the SOD1 dataset: use the custom script Rebuild_2D_spectra to reconstruct the 2D spectral regions from the 1D output of MCR-ALS and plot them.
7. Trypan blue test
- Recover the NMR tube content with a Pasteur pipet, and transfer the agarose threads to a 1.5 mL capped tube.
- Remove the residual medium by rinsing the agarose threads with 600 µL of PBS and centrifuge them at 4,000 x g for 1 min at room temperature. Discard the supernatant.
- Add 250 µL of PBS and 50 µL of 0.4% Trypan blue solution.
- Incubate for 2 min with continuous pipetting.
- Wash twice with 600 µL of PBS discarding the supernatant.
- Place a few agarose threads on a microscope slide and chop them with razor blades to create small slices of gel. Select the thinnest slices (thickness < 0.4 mm, ideally ~0.2 mm) for the analysis.
- Transfer the gel slices into a self-made cell counting chamber consisting of two glass slides spaced by three layers of paraffin film (~0.4 mm total thickness) on each side.
NOTE: A chamber slide could also be used; however, the gel slices thicker than the chamber height (0.1 mm) would be squeezed, rupturing the embedded cells. - Acquire images of cells inside the agarose and count white and blue cells.
- Calculate cell viability as (total cells - blue cells) / total cells.
Results
The above protocol allows the encapsulation of cells in threads of hydrogel to maximize cell viability for long periods of time, necessary to investigate in real time intracellular processes. In the bioreactor, the cells are maintained alive and metabolically active up to 72 h, as confirmed by Trypan blue test (Figure 2a-c). In principle, this protocol can be applied to observe an intracellular protein of interest undergoing any conformational or chemical changes. In the first application described above, the bioreactor is applied to monitor in real time the binding of two inhibitors, AAZ and MZA, to CA II overexpressed in the cytosol of HEK293T cells. The first 1H excitation sculpting spectrum (zgesgp) recorded is used to assess the overall signal intensity (which is proportional to the number of cells), the presence of signals from the overexpressed protein and the field homogeneity (Figure 2d). In the case of CA II, the intracellular binding of the two inhibitors can be monitored by WATERGATE 3-9-1935 1D 1H NMR spectra (p3919gp), by observing 1H signals in the region between 11 and 16 ppm. These signals arise from the zinc-coordinating histidines and other aromatic residues of CA II36 and are perturbed upon ligand binding14,15. Ligand concentrations can be chosen based on the diffusion rate or, if available, from previously determined permeability values14. Successful binding is confirmed visually by the appearance of an additional set of signals in the spectral region of interest, that gradually replaces the original signals (Figure 3). Time-dependent binding curves are obtained by MCR-ALS analysis, which separates the two sets of NMR signals arising from free and bound CA II (Figure 4a) and simultaneously provides the relative concentration profiles of the two species (Figure 4b). In the second application, the bioreactor is applied to monitor the formation of zinc-bound SOD1 intramolecular disulfide bond promoted by ebselen, a glutathione peroxidase mimetic, in human cells. This process is monitored by observing changes in 1H-15N 2D SOFAST-HMQC37 spectra (which provides a fingerprint of the protein backbone conformation) caused by perturbations of the protein structure induced by the disulfide bond formation. Additional signals arising from disulfide-oxidized SOD1 appear in the 1H-15N spectrum and gradually replace those from disulfide-reduced SOD1. MCR-ALS analysis on selected regions of the 2D spectrum separates the signals arising from the two species (Figure 5a) and provides their relative concentration profiles (Figure 5b). The obtained concentration curves can be further analyzed by non-linear fitting to provide information on the kinetics of the processes under study25.
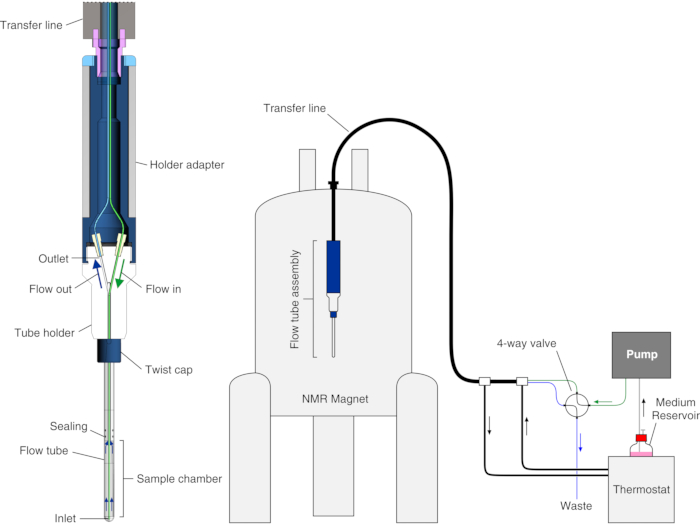
Figure 1: Scheme of the bioreactor. Left: cross-section view of the empty flow unit. Right: scheme of the bioreactor setup. The PEEK inlet tubing is shown in green; the PTFE outlet tubing is shown in blue. The left panel is reproduced with permission from Luchinat et al.25. Please click here to view a larger version of this figure.
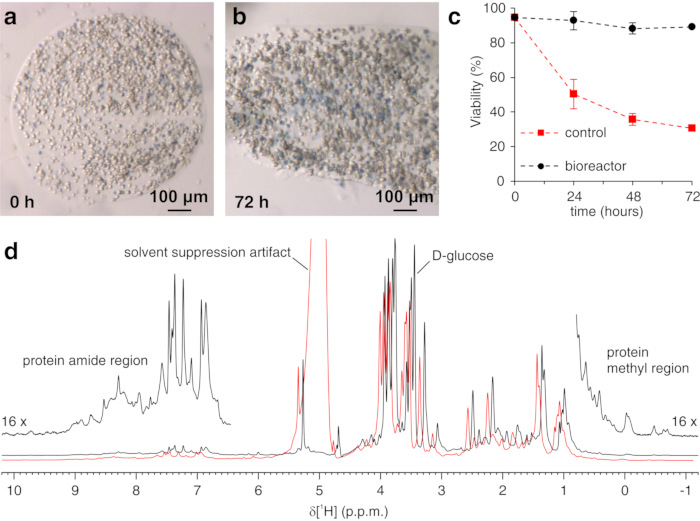
Figure 2: Trypan Blue Test on encapsulated cells and sample check by 1H NMR. Representative slices of agarose containing embedded cells and stained with Trypan blue (a) immediately after casting and (b) after 72 h in the bioreactor; (c) cell viability as a function of time in the NMR bioreactor under active flow (black) and under static conditions (red), measured by Trypan Blue Test. (d) zgesgp 1H NMR spectra recorded on agarose-embedded cells overexpressing CA II in the absence (black) and in the presence (red) of gas bubbles in the bioreactor. In the latter case, decreased field homogeneity causes line broadening and the appearance of solvent suppression artifacts. Interesting spectral features are labeled. Panels (a-c) are reproduced with permission from Luchinat et al.25. Please click here to view a larger version of this figure.
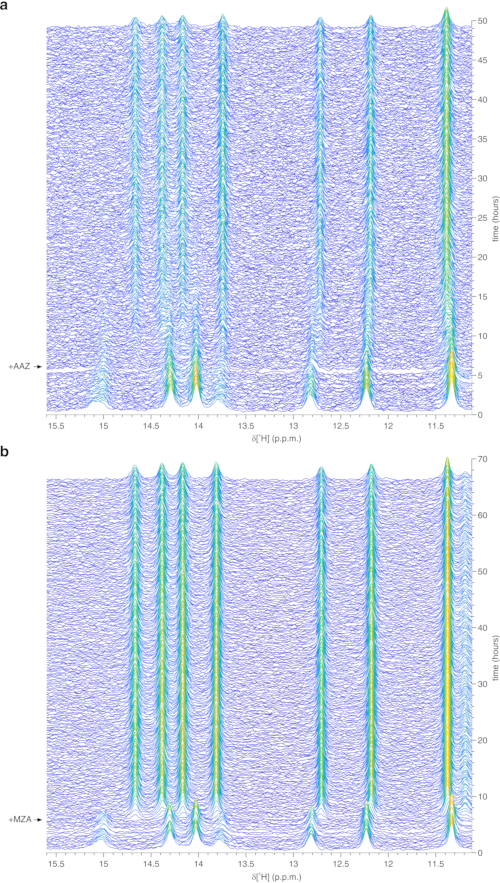
Figure 3: Representative real-time in-cell 1H NMR data obtained on agarose-encapsulated cells in the bioreactor. Waterfall plots of 1D 1H NMR spectra (region between 15.6 and 11.1 ppm) of HEK293T cells overexpressing CA II and subsequently treated with (a) 25 µM AAZ and (b) 10 µM MZA, recorded as a function of time in the NMR bioreactor. The time of ligand treatment is shown with an arrow. Spectral intensity (a.u.) is color-coded from blue (lowest) to yellow (highest). This figure is reproduced with permission from Luchinat et al.25. Please click here to view a larger version of this figure.
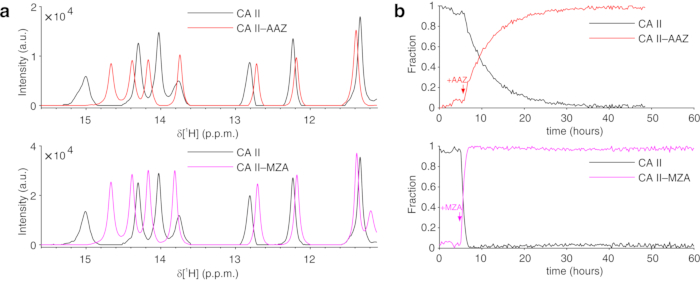
Figure 4: Representative MCR-ALS output from 1D NMR spectra. (a) 1H NMR spectra of the pure components reconstructed by MCR-ALS: CA II in the absence of ligands (black) and in the complex with AAZ (red) or MZA (magenta); (b) relative concentration profiles of free (black) and bound CA II as a function of time upon addition of AAZ (red) or MZA (magenta) obtained by MCR-ALS. Times of ligand treatment are marked with arrows. This figure is reproduced with permission from Luchinat et al.25. Please click here to view a larger version of this figure.
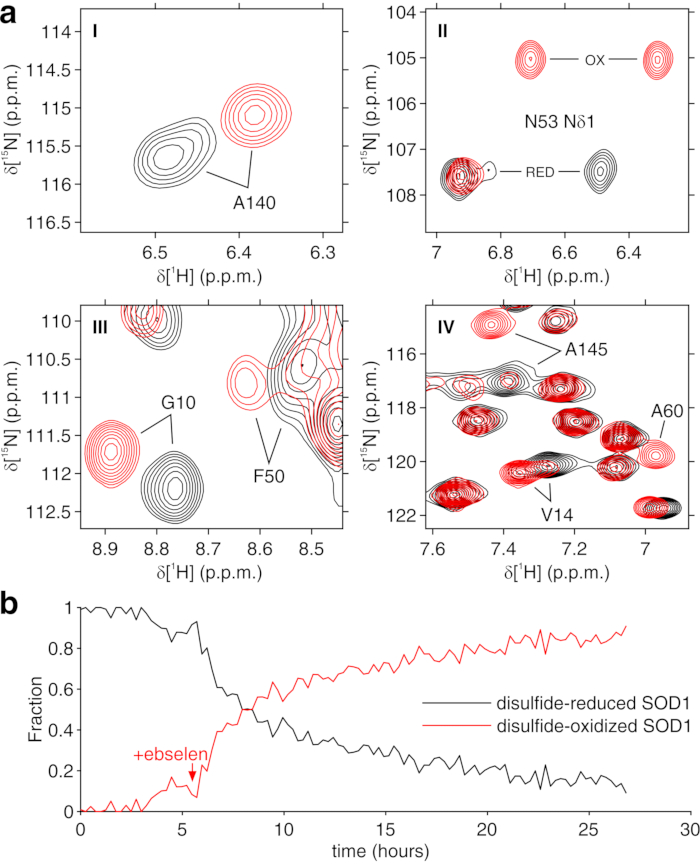
Figure 5: Representative MCR-ALS output from 2D NMR spectra. (a) 1H-15N spectral regions (labeled I-IV) of the pure components reconstructed by MCR-ALS: disulfide-reduced SOD1 (black) and disulfide-oxidized SOD1 (red); (b) relative concentration profile of the pure components as a function of time upon addition of ebselen (marked with an arrow) obtained by MCR-ALS. This figure is reproduced with permission from Luchinat et al.25. Please click here to view a larger version of this figure.
Discussion
The aim of using a bioreactor for in-cell NMR experiment is to keep cells alive and metabolically active for a prolonged period of time. A number of critical aspects must be taken into consideration to achieve this aim. First, it is paramount to avoid bacterial contamination when preparing the cell sample and during the NMR data acquisition. If strains of E. coli or other bacteria commonly used for gene cloning and recombinant protein expression are used in the laboratory, they may contaminate the cells during sample preparation. Once in the bioreactor, the bacteria will grow quickly exploiting the fresh growth medium and will cause cell death due to the production of endotoxins. Bacterial contamination is only spotted at an advanced stage, when it turns the growth medium yellow and turbid. Furthermore, incomplete cleaning of the bioreactor could cause contamination of the pump or the tubing with bacteria, yeasts, or common molds.
A requirement for the success of the experiment is the avoidance of gas bubble formation. Gas bubbles trapped between the agarose threads in the active volume of the NMR coil would introduce large magnetic field inhomogeneities, causing incomplete suppression of the H2O signal and severe loss of spectral quality (Figure 2d). Bubbles may be caused by air trapped in the system or by the formation of gaseous CO2. The former can be easily avoided by flushing the system with medium prior to inserting the cell sample, while to avoid the latter it is recommended to decrease the concentration of NaHCO3 in the growth medium, and to keep all parts of the system at a constant temperature to minimize differences in the CO2 solubility. Cellular aerobic metabolism may also cause the formation of gaseous CO2, which can be prevented by increasing the flow rate.
Cell viability should be checked after each run by Trypan Blue Test. However, that does not provide insights on the metabolic activity. To obtain a more complete picture of the metabolic state of the cells during the bioreactor operation, 31P NMR spectra can be performed to assess the production of ATP as a function of time23,25. However, a dedicated probe is often necessary for this measurement, which may allow simultaneous recording with 1H NMR.
In the case of CA II, the presence of well-resolved reporter signals in an unusual region of the 1H spectrum facilitates the analysis from simple 1D NMR spectra and does not require isotope enrichment during protein expression. In general, other proteins could give rise to 1H signals useful for monitoring spectral changes in other regions, such as that typical of the protein hydrophobic core between 0 and -1 ppm11; however, these regions tend to be crowded for folded proteins larger than ~10 kDa. In this case, as shown for SOD1, it is preferable to enrich the protein with 15N, by providing uniformly 15N-enriched growth medium during protein expression, and to monitor real-time changes in 2D 1H-15N NMR spectra. 2D spectra are imported as a 2D arrays in MATLAB, rearranged to 1D arrays and stacked prior to MCR-ALS analysis. The latter approach is generally applicable to any intracellular protein that gives rise to detectable signals, and provides information on protein conformational changes at the single residue level. In principle, the latter approach can be generalized to nD spectra and to other isotope-labeling schemes.
Concerning the application to different types of cells, the protocol should be easily adapted to different cell lines and does not require that the protein of interest is directly expressed in the cells. Therefore, other approaches to in-cell NMR can be combined with this protocol, in which the macromolecule of interest is produced recombinantly, or synthesized, and subsequently inserted into the cells by electroporation or by other delivery methods1,9,38. When working with different cell lines or sample preparation protocols, parameters such as the agarose concentration, the thread thickness, and the final cell density in the agarose threads may need to be optimized empirically. Furthermore, the applicability of the protocol described here is limited to cells that tolerate agarose encapsulation. Other cell types may require different hydrogel formulations, whereas a different setup is recommended when analyzing cells that natively grow in suspension, for example, making use of a coaxial microdialysis membrane to ensure nutrient diffusion while keeping suspended cells confined in the NMR tube23.
Compared to other NMR bioreactor designs19,20,21,22, the device described here relies on a commercially available flow-unit, adapted with minor modifications. Therefore, the device can be easily replicated in different laboratories with high reproducibility. Furthermore, it allows standardized operation and full compliance with strict laboratory safety regulations, if needed. Overall, the flexibility and ease of operation of the bioreactor should allow many other applications of solution NMR, both in cells and in vitro, in addition to those as already reported23,25. Eventually, the same bioreactor design could be applied to samples that resemble more of the physiological environment of a tissue, such as spheroids or organoids, provided that appropriate scaffolds are found for keeping such samples alive-or even sustaining their growth-in the NMR spectrometer.
Disclosures
The authors declare that there is no conflict of interest.
Acknowledgements
This work has been supported by iNEXT-Discovery, grant number 871037, funded by the Horizon 2020 programme of the European Commission, by Instruct-ULTRA, grant number 731005, an EU H2020 project to further develop the services of Instruct-ERIC, and by Ministero dell'Istruzione, dell'Università e della Ricerca PRIN grant 20177XJCHX. The authors acknowledge the support of Instruct-ERIC, a Landmark ESFRI project, through the JRA Award number 815 and the use of resources of the CERM/CIRMMP Italy Centre. We thank Matteo Pennestri (Bruker, UK) for providing support for the InsightMR flow unit operation.
Materials
| Name | Company | Catalog Number | Comments |
| Materials | |||
| Citric acid | Sigma-Aldrich | 251275 | |
| D2O | Sigma-Aldrich | 453366 | |
| DMEM, high glucose | Life Technologies | 10313-021 | |
| DMEM, high glucose, powder | Sigma-Aldrich | D5648 | |
| FBS | Life Technologies | 10270 | |
| HCl | Sigma-Aldrich | 30721 | |
| L-glutamine (200 mM) | Life Technologies | 25030 | |
| Low-gelling agarose, powder | Sigma-Aldrich | A4018 | |
| NaHCO3, powder | Carlo Erba | 478537 | |
| PBS | Life Technologies | 10010 | |
| Penicillin–streptomycin (10,000 U/ml) | Life Technologies | 15140-122 | |
| NaOH, pellets | Sigma-Aldrich | 30620 | |
| Trypan Blue solution (0.4% (wt/vol) | Sigma-Aldrich | T8154 | Hazard statement(s): H350 may cause cancer. |
| Trypsin–EDTA (0.05% (wt/vol)) | Life Technologies | 25300-054 | |
| Equipment | |||
| Avance III Spectrometer equipped with a 5 mm CryoProbe | Bruker | n/a | All modern spectrometers and narrow-bore magnets equipped with 5 mm probes are compatible. |
| InsightMR flow unit | Bruker | n/a | |
| P-920 pump module from ÄKTA FPLC | GE Healthcare | n/a | Any FPLC, HPLC peristaltic or syringe pump should be compatible with the flow unit. |
References
- Inomata, K., et al. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature. 458 (7234), 106-109 (2009).
- Luchinat, E., Banci, L. In-cell NMR: a topical review. IUCrJ. 4, 108-118 (2017).
- Dzatko, S., et al. Evaluation of the stability of DNA i-motifs in the nuclei of living mammalian cells. Angewandte Chemie (International Ed. in English). 57 (8), 2165-2169 (2018).
- Luchinat, E., Banci, L. In-cell NMR in human cells: direct protein expression allows structural studies of protein folding and maturation. Accounts of Chemical Research. 51 (6), 1550-1557 (2018).
- Tanaka, T., et al. High-resolution protein 3D structure determination in living eukaryotic cells. Angewandte Chemie (International Ed. in English). 58 (22), 7284-7288 (2019).
- Siegal, G., Selenko, P. Cells, drugs and NMR. Journal of Magnetic Resonance. 306, 202-212 (1997).
- Banci, L., et al. Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nature Chemical Biology. 9 (5), 297-299 (2013).
- Luchinat, E., et al. In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nature Communications. 5, 5502 (2014).
- Theillet, F. -. X., et al. Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature. 530 (7588), 45-50 (2016).
- Barbieri, L., Luchinat, E., Banci, L. Intracellular metal binding and redox behavior of human DJ-1. Journal of biological inorganic chemistry: JBIC: A Publication of the Society of Biological Inorganic Chemistry. 23 (1), 61-69 (2018).
- Banci, L., Barbieri, L., Luchinat, E., Secci, E. Visualization of redox-controlled protein fold in living cells. Chemistry & Biology. 20 (6), 747-752 (2013).
- Mercatelli, E., Barbieri, L., Luchinat, E., Banci, L. Direct structural evidence of protein redox regulation obtained by in-cell NMR. Biochimica Et Biophysica Acta. 1863 (2), 198-204 (2016).
- Barbieri, L., Luchinat, E., Banci, L. Protein interaction patterns in different cellular environments are revealed by in-cell NMR. Scientific Reports. 5, 14456 (2015).
- Luchinat, E., et al. Drug screening in human cells by NMR spectroscopy allows the early assessment of drug potency. Angewandte Chemie (International Ed. in English). 59 (16), 6535-6539 (2020).
- Luchinat, E., et al. Intracellular binding/unbinding kinetics of approved drugs to carbonic anhydrase II observed by in-cell NMR. ACS Chemical Biology. 15 (10), 2792-2800 (2020).
- DeMott, C. M., et al. Potent inhibitors of mycobacterium tuberculosis growth identified by using in-cell NMR-based screening. ACS Chemical Biology. 13 (3), 733-741 (2018).
- Krafcikova, M., et al. Monitoring DNA-ligand interactions in living human cells using NMR spectroscopy. Journal of the American Chemical Society. 141 (34), 13281-13285 (2019).
- Broft, P., et al. In-cell NMR of functional riboswitch aptamers in eukaryotic cells. Angewandte Chemie (International Ed. in English). , (2020).
- Sharaf, N. G., Barnes, C. O., Charlton, L. M., Young, G. B., Pielak, G. J. A bioreactor for in-cell protein NMR. Journal of magnetic resonance. 202 (2), 140-146 (2010).
- Kubo, S., et al. A gel-encapsulated bioreactor system for NMR studies of protein-protein interactions in living mammalian cells. Angewandte Chemie (International Ed. in English). 52 (4), 1208-1211 (2013).
- Inomata, K., Kamoshida, H., Ikari, M., Ito, Y., Kigawa, T. Impact of cellular health conditions on the protein folding state in mammalian cells. Chemical Communications. 53 (81), 11245-11248 (2017).
- Breindel, L., DeMott, C., Burz, D. S., Shekhtman, A. Real-time in-cell nuclear magnetic resonance: ribosome-targeted antibiotics modulate quinary protein interactions. Biochemistry. 57 (5), 540-546 (2018).
- Cerofolini, L., et al. Real-time insights into biological events: in-cell processes and protein-ligand interactions. Biophysical Journal. 116 (2), 239-247 (2019).
- Breindel, L., Burz, D. S., Shekhtman, A. Active metabolism unmasks functional protein-protein interactions in real time in-cell NMR. Communications Biology. 3, (2020).
- Luchinat, E., Barbieri, L., Campbell, T. F., Banci, L. Real-time quantitative in-cell NMR: ligand binding and protein oxidation monitored in human cells using multivariate curve resolution. Analytical Chemistry. 92 (14), 9997-10006 (2020).
- Koczula, K. M., et al. Metabolic plasticity in CLL: adaptation to the hypoxic niche. Leukemia. 30 (1), 65-73 (2016).
- Alshamleh, I., et al. Real-time NMR spectroscopy for studying metabolism. Angewandte Chemie (International Ed. in English). 59 (6), 2304-2308 (2020).
- Supuran, C. T. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nature Reviews. Drug Discovery. 7 (2), 168-181 (2008).
- Neri, D., Supuran, C. T. Interfering with pH regulation in tumours as a therapeutic strategy. Nature Reviews. Drug Discovery. 10 (10), 767-777 (2011).
- Alterio, V., Di Fiore, A., D'Ambrosio, K., Supuran, C. T., De Simone, G. Multiple binding modes of inhibitors to carbonic anhydrases: how to design specific drugs targeting 15 different isoforms. Chemical Reviews. 112 (8), 4421-4468 (2012).
- Capper, M. J., et al. The cysteine-reactive small molecule ebselen facilitates effective SOD1 maturation. Nature Communications. 9 (1), 1693 (2018).
- Trist, B., Hilton, J. B., Crouch, P. J., Hare, D. J., Double, K. L. Superoxide dismutase 1 in health and disease: How a front-line antioxidant becomes neurotoxic. Angewandte Chemie (International Ed. in English). , (2020).
- Tauler, R. Multivariate curve resolution applied to second order data. Chemometrics and Intelligent Laboratory Systems. 30 (1), 133-146 (1995).
- Barbieri, L., Luchinat, E., Banci, L. Characterization of proteins by in-cell NMR spectroscopy in cultured mammalian cells. Nature Protocols. 11 (6), 1101-1111 (2016).
- Piotto, M., Saudek, V., Sklenár, V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. Journal of biomolecular NMR. 2 (6), 661-665 (1992).
- Vasa, S. K., Singh, H., Grohe, K., Linser, R. Assessment of a large enzyme-drug complex by proton-detected solid-state NMR spectroscopy without deuteration. Angewandte Chemie (International Ed. in English). 58 (17), 5758-5762 (2019).
- Schanda, P., Brutscher, B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. Journal of the American Chemical Society. 127 (22), 8014-8015 (2005).
- Ogino, S., et al. Observation of NMR signals from proteins introduced into living mammalian cells by reversible membrane permeabilization using a pore-forming toxin, streptolysin O. Journal of the American Chemical Society. 131 (31), 10834-10835 (2009).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved