A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
RIBO-seq in Bacteria: a Sample Collection and Library Preparation Protocol for NGS Sequencing
In This Article
Summary
Here we describe the stages of sample collection and preparation for RIBO-seq in bacteria. Sequencing of the libraries prepared according to these guidelines results in sufficient data for comprehensive bioinformatic analysis. The protocol we present is simple, uses standard laboratory equipment and takes seven days from lysis to obtaining libraries.
Abstract
The ribosome profiling technique (RIBO-seq) is currently the most effective tool for studying the process of protein synthesis in vivo. The advantage of this method, in comparison to other approaches, is its ability to monitor translation by precisely mapping the position and number of ribosomes on a mRNA transcript.
In this article, we describe the consecutive stages of sample collection and preparation for RIBO-seq method in bacteria, highlighting the details relevant to the planning and execution of the experiment.
Since the RIBO-seq relies on intact ribosomes and related mRNAs, the key step is rapid inhibition of translation and adequate disintegration of cells. Thus, we suggest filtration and flash-freezing in liquid nitrogen for cell harvesting with an optional pretreatment with chloramphenicol to arrest translation in bacteria. For the disintegration, we propose grinding frozen cells with mortar and pestle in the presence of aluminum oxide to mechanically disrupt the cell wall. In this protocol, sucrose cushion or a sucrose gradient ultracentrifugation for monosome purification is not required. Instead, mRNA separation using polyacrylamide gel electrophoresis (PAGE) followed by the ribosomal footprint excision (28-30 nt band) is applied and provides satisfactory results. This largely simplifies the method as well as reduces the time and equipment requirements for the procedure. For library preparation, we recommend using the commercially available small RNA kit for Illumina sequencing from New England Biolabs, following manufacturer's guidelines with some degree of optimization.
The resulting cDNA libraries present appropriate quantity and quality required for next generation sequencing (NGS). Sequencing of the libraries prepared according to the described protocol results in 2 to 10 mln uniquely mapped reads per sample providing sufficient data for comprehensive bioinformatic analysis. The protocol we present is quick and relatively easy and can be performed with standard laboratory equipment.
Introduction
The ribosome profiling technique (RIBO-seq) was developed in the laboratory of Jonathan Weissman at the University of California, San Francisco1. In comparison to other methods used to study gene expression at the translational level, RIBO-seq focuses on each ribosome binding to mRNA and provides information about its location and the relative number of ribosomes on a transcript. It enables monitoring the process of protein synthesis in vivo and can provide single codon resolution and accuracy allowing the measurement of the ribosome density on both, the individual mRNA and along the entire transcriptome in the cell. At the foundation of the RIBO-seq technique lies the fact that during translation the ribosome binds the mRNA molecule and thus protects the buried fragment of the transcript from a ribonuclease digestion. Upon addition of the ribonuclease, the unprotected mRNA is digested and the fragments enclosed by ribosomes - typically of ~28-30 nt long - remain intact. These fragments, called ribosomal footprints (RF), can then be isolated, sequenced and mapped onto the transcript they originated from resulting in the detection of the exact position of the ribosomes. In fact, the ribosome ability to protect mRNA fragments has been used since the 1960s to study ribosomal binding and translation initiation sites (TIS)2,3,4. However, with the advancement in deep sequencing technology, RIBO-seq has become a gold standard for translation monitoring5 which, through the ribosome engagement, can provide a genome-wide information on protein synthesis6. Ribosome profiling filled the technological gap that existed between quantifying the transcriptome and the proteome6.
To conduct ribosome profiling we need to obtain cell lysate of the organism that had grown under the investigated conditions. Disrupting these conditions during cell collection and lysis may provide unreliable data. To prevent this, translation inhibitors, rapid harvesting and flash freezing in liquid nitrogen are commonly used. Cells can be lysed by cryogenic grinding in a mechanical homogenizer like a mixer mill7,8 or a bead beater9, and by trituration through a pipette10 or with a needle11. The lysis buffer can be added just before or shortly after pulverization of the cells. In our protocol we use liquid nitrogen to precool mortar and pestle, as well as aluminum oxide as a gentler approach to disruption of the bacterial cell wall, which prevents RNA shearing often encountered when methods such as sonification are applied. After pulverization, we add an ice-cold lysis buffer into the cooled contents of the mortar. Selection of an appropriate lysis buffer is important for obtaining the best resolution of ribosomal footprints. Since ionic strength affects both the RF size and the reading frame precision, it is currently recommended to use lysis buffers with low ionic strength and buffer capacity, even if it appears that buffer composition does not affect ribosomal occupancy on mRNAs11,12. Important components of the lysis buffer are magnesium ions, the presence of which prevents dissociation of the ribosomal subunits and inhibits spontaneous conformational changes in the bacterial ribosomes11,13. Calcium ions also play a significant role and are essential for the activity of micrococcal nuclease (MNase) used in the bacterial ribosome profiling method14. Addition of guanosine 5′-[β,γ-imido]triphosphate (GMP-PNP), a non-hydrolyzable analog of GTP, together with chloramphenicol inhibits translation during lysis15.
When the lysate is obtained, it is clarified by centrifugation and divided into two portions, each for a RIBO-seq and a high-throughput total mRNA sequencing (RNA-seq) since they are performed simultaneously (Figure 1). RNA-seq provides a point of reference which enables the comparison of data from both RIBO-seq and RNA-seq during data analysis. The investigated translatome is defined by normalization of ribosomal footprints to mRNA abundance16. Data from RNA-seq can also help identify cloning or sequencing artifacts17.
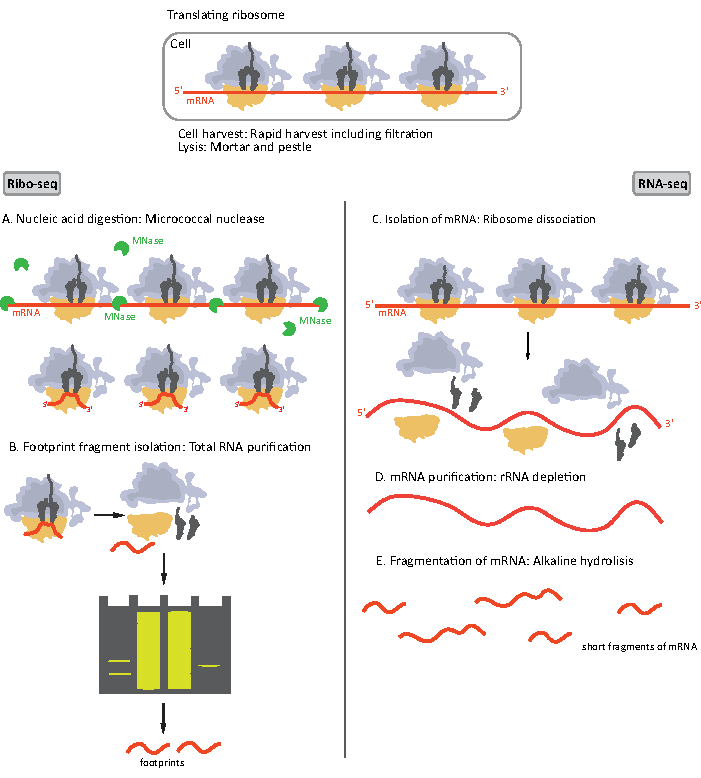
Figure 1. Schematics of mRNA sample preparation for RIBO-seq and RNA-seq. For RIBO-seq library preparation, RNA is digested with MNase (A), followed by the size selection of RF of ~28-30 nt length (B); for RNA-seq RNA is isolated (C), depleted of rRNA (D), and the resulting mRNA is randomly fragmented into fragments of varying lengths (E). Please click here to view a larger version of this figure.
Initial steps of the procedure of sample preparation for RIBO-seq and RNA-seq differ slightly (Figure 1). For the ribosomal profiling, the lysate needs to be digested by a specific endonuclease to degrade the mRNA molecules not protected by the ribosomes. In standard protocols, the obtained monosomes are recovered by a sucrose cushion ultracentrifugation or a sucrose gradient ultracentrifugation8,14. In this article, we show that this step is not necessary to isolate RF required for the RIBO-seq in bacteria, likewise for eukaryotic cells18, and that size selection of the appropriate length mRNA fragments from the polyacrylamide gel is sufficient.
For RNA-seq, mRNA is obtained by the depletion of rRNA from the total RNA - rRNA molecules hybridize to the biotinylated oligonucleotide probes which bind to the streptavidin-coated magnetic beads. The rRNA-oligonucleotide-bead complexes are then removed from the sample with a magnet resulting in a rRNA depleted sample19,20. The purified mRNA molecules are then randomly fragmented by alkaline hydrolysis. The obtained fragments of mRNA as well as the ribosomal footprints are converted into cDNA libraries and prepared for deep sequencing (Figure 2). This involves ends repair needed after alkaline hydrolysis (for mRNA) and enzymatic digestion (for RF): dephosphorylation of 3' ends followed by phosphorylation of 5' ends. The next steps are adaptors ligation and the reverse transcription to create cDNA inserts framed by sequences required for the next generation sequencing (NGS) using Illumina platform. The last phase of library preparation is a PCR reaction in which the constructs are amplified and labelled with sample specific barcodes to allow multiplexing and sequencing various samples on one channel. Before sequencing, the quality and quantity of the libraries are assessed by the high-sensitivity DNA on-chip electrophoresis. cDNA libraries with appropriate parameters can then be pooled and sequenced. Sequencing can be performed on different Illumina platforms, such as MiSeq, NextSeq or HighSeq, depending on the number of libraries, required read length and sequencing depth. After sequencing, the bioinformatic analysis is performed.
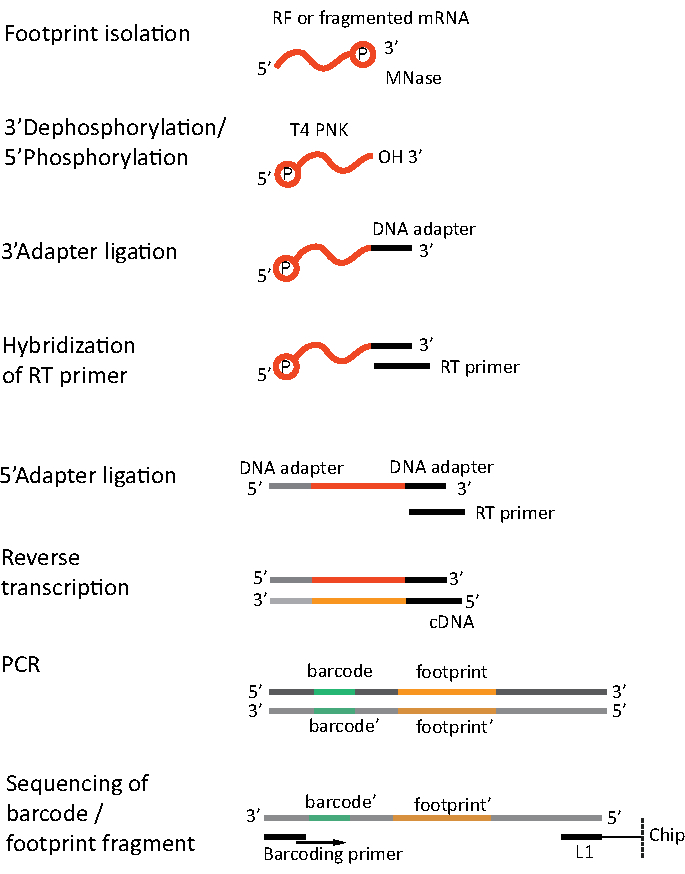
Figure 2. Library preparation. Library preparation includes the ends repair, adapters ligation, reverse transcription and amplification with barcoding. Please click here to view a larger version of this figure.
The ribosome profiling is a universal method which can be easily modified and adjusted according to the scientific question. Originally it was used in yeast1, but shortly after it was applied to bacterial cells21 as well as eukaryotic model organisms including mouse10, zebrafish22, fruit fly23 and Arabidopsis thaliana24. It was also used for studying different ribosome types: cytoplasmic, mitochondrial25,26 and chloroplast27,28. In eukaryotes RIBO-seq is commonly adapted and refined to investigate specific aspects of translation, including initiation10,11,29,30,31,32, elongation1,10,11,31,33, ribosome stalling33 and conformation change33. Most of the modifications involve the use of different translation inhibitors. In bacteria however, analogous studies have been difficult to conduct because of the paucity of inhibitors with the required mechanism of action34. The most commonly used translation inhibitor in bacteria is chloramphenicol (CAM) which binds to the peptidyl transferase center (PTC) and prevents correct positioning of the aminoacyl-tRNA in the A-site. As a result, CAM prevents the formation of a peptide bond which leads to arresting the elongating ribosomes35. Other examples of translation inhibitors in bacteria are tetracycline (TET)36, retapamulin (RET)34 and Onc11237 which have been used to investigate translation initiation sites. TET, which prevents tRNA delivery to the ribosome by directly overlapping with the anticodon stem-loop of tRNA at the A-site, was originally applied to verify the results obtained from CAM treatment since they are both antibiotics inhibiting translation elongation38. TET was found to detect primary TIS, however was unable to reveal internal TIS36. RET binds in the PTC of the bacterial ribosome, and prevents formation of the first peptide bond by interfering with an elongator aminoacyl-tRNA in the A site. Applying RET results in ribosomes arrest at both primary as well as internal TISs34. Onc112, a proline-rich antimicrobial peptide, binds in the exit tunnel and blocks aminoacyl-tRNA binding in the ribosomal A site. As a result, Onc112 prevents initiation complexes from entering the elongation phase37.
The main information ribosome profiling provides is ribosomes density and their position on the mRNA. It was successfully applied to investigate differential gene expression at the level of translation in various growth conditions1,6, measure translational efficiency1,38,39 and detect translation regulation events such as ribosomal pausing10. RIBO-seq also allows for uncovering the translation of annotated ncRNA, pseudogenes and unannotated small open reading frames (ORF) leading to the identification of novel and/or very short protein coding genes10,12,22,30,37. In such cases, RIBO-seq can fine-tune and improve genome annotation. With its high sensitivity for the identification of translated ORFs and its quantitative nature, ribosome profiling can also serve as a proxy for the proteome determination or in aiding proteomics studies31,34,39. By mapping TIS, ribosome profiling reveals N-terminally extended and truncated isoforms of known proteins10,32. RIBO-seq was also adapted to study co-translational folding of proteins14,21,24. This method enables measuring of elongation rates1,10,39 or decoding speeds of individual codons6 and helps in developing quantitative models of translation17. The ribosome profiling method is also capable of providing mechanistic insights into the ribosome pausing in bacteria7,15,17, frameshifting40, stop-codon readthrough21, termination/recycling defects41,42 and ribosomal conformation changes33 in eukaryotes. RIBO-seq was also adapted to examine the impact of specific trans-acting factors on translation such as miRNAs6 and RNA-binding proteins in eukaryotes16,43. However, it is important to acknowledge that the experimental design and the obtained resolution of RIBO-seq determine the amount of information that can be extracted from the resulting sequencing data12.
Protocol
1. Sample collection
- Prepare a bacterial culture. We recommend a culture volume of 100 mL per sample.
- Prepare equipment and reagents for sample collection: two scoopulas per sample, sterile 0.45 µm mixed cellulose esters membrane (MCE) filters, 50 mL sterile tubes, liquid nitrogen, 50 mg/mL chloramphenicol in 70% (vol/vol) ethanol (optional). Puncture the lid of 50 mL tubes to allow liquid nitrogen evaporation and prevent the explosion of closed tubes. Decontaminate scoopulas with laboratory detergent and 70% ethanol.
- Prewarm the filtering equipment and one of the scoopulas to growth temperature of the bacterial culture.
NOTE: We recommend an all-glass filtration apparatus with a funnel, a fritted base, a ground joint flask and a 47 mm Ø spring clamp. - Before sample harvesting, add chloramphenicol into the bacterial culture to the final concentration of 100 µg/mL and incubate 1 minute (optional).
- Pour liquid nitrogen into 50 mL sterile tube and place the second scoopula inside the tube to cool.
CAUTION! Liquid nitrogen can cause closed containers to explode due to the pressure change upon evaporation. To prevent this, it is necessary to create a few holes in the lid of 50 mL tubes to allow liquid nitrogen evaporation. - Collect cells by filtration. Stop filtering when medium has passed through the membrane, but do not allow the filter to dry completely.
- Collect bacterial pellets by rapidly scraping the cells off of the filter disc using a prewarmed scoopula. Immediately place the entire scoopula with the harvested cells in the 50 mL tube filled with liquid nitrogen. The harvested pellet should be completely covered in liquid nitrogen.
- Let the pellet freeze thoroughly and dislodge frozen cells using a previously cooled second scoopula. Close the punctured lid and transfer the tube to -80°C. STOP point
NOTE: Disinfect scoopulas after each sample harvesting.
2. Cell lysis
- Prepare 0.1 M GMP-PNP in 10 mM Tris pH 8, DNase I and 1.5 mL reaction tubes for lysates and keep them on ice. Cool the centrifuge to 4 °C.
- Prepare lysis buffer (20 mM TRIS pH 7.6, 10 mM MgAcet, 150 mM KAcet, 0.4% TRITON X-100, 6 mM β-mercaptoethanol, 5 mM CaCl2 and optional 1 mM chloramphenicol). Aliquot 500 µL of the lysis buffer per sample into 1.5 mL reaction tubes and place them on ice.
- Decontaminate mortar and pestle with a laboratory detergent and 70% ethanol, and dry them. Cool mortar and pestle by pouring liquid nitrogen into the mortar.
- Transfer frozen pellet into the pre-chilled mortar and grind it to a powder. With a spatula, add approximately 1 volume of aluminum oxide and continue grinding. Keep mortar, pestle and cells cool by pouring liquid nitrogen when needed, do not let the contents of the mortar to thaw.
- Just before using lysis buffer, add GMP-PNP and DNase I into the lysis buffer aliquot to the final concentration of 2 mM and 100 U/mL respectively. Transfer the solution into the mortar with cells and aluminum oxide and continue grinding. Let the lysate thaw slowly while grinding and transfer the mixture into the pre-cooled 1.5 mL reaction tube and immediately place on ice.
- Centrifuge lysates at 20 000 x g for 5 minutes at 4 °C. Transfer supernatants into new, pre-cooled 1,5 mL reaction tubes and keep them on ice.
- Measure the RNA concentration in each sample with a NanoDrop. Use 1:10 dilutions of samples and 1:10 dilution of lysis buffer in nuclease-free water as a blank.
NOTE: Be aware that chloramphenicol shows significant absorption at 260 nm. - Divide each lysate into two portions: one for RIBO-seq (0.5 - 1 mg of RNA) and the second for RNA-seq (the rest).
- Clean the samples for RNA-seq using a commercially available RNA clean-up kit according to the manufacturer's protocol.
NOTE: We recommend using Zymo RNA Clean & Concentrate -25 kit (see Table of Materials). We also recommend adding 4.5 volumes of ethanol (compared to sample volume) into the mix of sample and Binding Buffer for ribosomal footprints and fragmented mRNA, in order to increase the purification efficiency of short RNA fragments. - Store the samples intended for RNA-seq at -80 °C. The procedure for RNA-seq will continue from step 5.
3. MNase digestion of samples for RIBO-seq
- To 1 mg of RNA add 3.8 µL of 187.5 U/µL MNase in 10 mM Tris pH 8 and top it up with lysis buffer to the total volume of 500 µL.
- Incubate at 25 °C, 300 RPM, for 45 minutes in a thermomixer.
- Clean the samples with a commercially available RNA clean up kit (as in step 2.9).
- Store the samples for RIBO-seq at -80 °C (STOP point) or proceed to size selection.
4. Polyacrylamide gel electrophoresis (PAGE) and size selection of samples for RIBO-seq
- Prepare a 15% polyacrylamide-TBE gel with 8 M urea and place it in a tank with TBE buffer. Pre-run for minimum 10 minutes at a constant voltage of 200 V.
- Mix samples with TBE-Urea Sample Buffer, denature at 95 °C for 1 minute and place them immediately on ice.
- Wash out the urea by injecting TBE buffer into gel wells using a syringe. Load the samples leaving one well space between them to separate each sample and prevent cross-contamination. Use 29 nt oligonucleotide and a mix of 26 nt and 32 nt oligonucleotides as markers. Run electrophoresis at a constant voltage of 180 V.
- Prepare sterile buffer for overnight incubation (5 mM EDTA, 10 mM NaOAc pH 5).
- After electrophoresis, stain the gel in a SYBR Gold-bath for about 2-3 minutes and rinse the gel with nuclease-free water.
- Excise fragments of gel between 26 and 32 nt with the sterile needles or the razor blades and place the gel fragments in separate 1.5 mL tubes for each sample. Change the needle or the razor blade between the samples.
- Add 200 µL of the overnight incubation buffer to each reaction tube.
- Incubate the samples at 10 °C, 1000 RPM overnight in a thermomixer.
- The next day, clean the samples as in step 2.9. Elute the footprints in 80 µL of nuclease-free water.
- Store the samples at -80 °C. STOP point. The procedure for RIBO-seq will continue from step 7.
5. Purification of bacterial mRNA by rRNA depletion from samples for RNA-seq
- Remove rRNA from samples with a bacterial mRNA purification kit (e.g., Invitrogen MICROBExpress). Follow the manufacturer's protocol.
- Clean the samples as in step 2.9. Elute the mRNA in 50 µL of nuclease-free water.
- Store the samples for RNA-seq at -80 °C (STOP point) or continue to alkaline fragmentation.
6. Alkaline fragmentation of samples for RNA-seq
- Prepare alkaline hydrolysis buffer (mix 220 µL of 0.1 M NaHCO3, 30 µL of 0.1 M Na2CO3 and 1 µL of 0.5 M EDTA). Mix 1 volume (50 µL) of alkaline buffer with 1 volume (50 µL) of the sample and incubate at 95 °C for 25 minutes.
- Add 5 µL of 3 M NaOAc pH 5.5 to stop the reaction.
- Clean the samples as in step 2.9 and elute the samples in 80 µL of nuclease-free water.
- Possible STOP point: store RNA-seq samples at -80 °C. However, we recommend to go directly to step 7 in order to avoid unnecessary freezing and thawing.
7. Dephosphorylation and phosphorylation of samples for both RNA- and RIBO-seq
- Add 10 µL of 10x reaction buffer PNK and 5 µL T4 PNK to each sample. Incubate at 37 °C for 1.5 hour in order to dephosphorylate the 3' ends.
- Add 3 µL of 1 mM ATP and incubate at 37 °C for 1 hour in order to phosphorylate the 5' ends.
- Clean the samples as in step 2.9, elute in 30 µL nuclease-free water.
- Store the samples at -80 °C. STOP point.
8. Library preparation using NEBNext Multiplex Small RNA Library Prep Set for Illumina
- Prepare 100-1000 ng of input RNA (obtained mRNA fragments and ribosomal footprints) in 6 µL of nuclease-free water and add 1 µL of 3' SR Adaptor. Incubate according to the manufacturer's protocol.
- Add 10 µL of 3' Ligation Reaction Buffer (2X) and 3 µL of 3' Ligation Enzyme Mix and incubate at 37 °C for 2.5 hour, instead of 1 hour at 25 °C as the manufacturer's protocol specifies.
- Hybridize the Reverse Transcription Primer according to the manufacturer's protocol with a modified program: 5 minutes at 75 °C, 30 minutes at 37 °C and hold at 4 °C.
- Ligate the 5´ SR Adaptor. Follow the manufacturer's protocol with one change: perform the incubation at 37 °C for 2.5 hour, instead of 1 hour at 25 °C.
- Perform Reverse Transcription according to the manufacturer's protocol.
- Possible STOP point: incubate the samples containing synthesized cDNAs at 70 °C for 15 minutes to inactivate the reverse transcriptase and store them at -80 °C. However, we recommend to proceed directly to the next steps to avoid unnecessary freezing and thawing of the samples.
- Store half of each cDNA library at -80 °C as a back-up.
- Perform PCR amplification according to the manufacturer's protocol. Use half of the reaction mix. Store the obtained libraries at -80 °C. STOP point.
9. Size selection of cDNA libraries using PAGE
- Prepare 6% polyacrylamide-TBE gel and place it in a tank with TBE buffer. Pre-run for minimum 10 minutes at a constant voltage of 200 V.
- Mix the samples with Gel Loading Dye, Blue (6X) and load them on the gel. Use Quick-Load pBR322 DNA-MspI Digest as a ladder. At the beginning, run electrophoresis at a constant voltage of 120 V to allow DNA to enter the gel and then change the voltage to 180 V.
- When electrophoresis is finished, stain the gel in a SYBR Gold-bath for about 2-3 minutes and rinse the gel with nuclease-free water.
- Excise gel fragments containing the libraries. For RNA-seq samples excise between 135-180 nt and for RIBO-seq between 135-170 nt. Use sterile needle or razor blade and place the excised gel fragments in separate 1.5 mL reaction tubes. Remember to change the needle or razor blade between the samples.
NOTE: The adapters and barcode from NEBNext Multiplex Small RNA Library Prep Set for Illumina have 119 nt in total which determines the lower excision cut-off. - Add 100 µL of nuclease-free water to each excised gel fragment.
- Incubate the gel fragments at 10 °C, 450 RPM overnight in a thermomixer.
- Clean and concentrate the cDNA libraries with a commercial DNA clean-up kit according to the manufacturer's protocol.
NOTE: We recommend using Zymo DNA Clean & Concentrate -5 kit (see Table of Materials). - Store purified cDNA libraries at -80 °C. STOP point.
Results
The exemplary results presented here were obtained in a study examining translation regulation in sporulating WT Bacillus subtilis cells. Overnight cultures were diluted to OD600 equal to 0.1 in 100 mL of rich medium and incubated at 37 °C with vigorous shaking until OD600 reached 0.5-0.6. The rich medium was then replaced with minimal medium to induce sporulation process and the incubation was continued for up to four hours. Cells were harvested every hour beginning with T0 - sporulat...
Discussion
The key technical challenge of the ribosome profiling is the need to rapidly inhibit translation in order to capture a snapshot of ribosomes on mRNAs at a particular physiological state of interest. To accomplish this, translation inhibitors, rapid harvesting and flash freezing in liquid nitrogen are commonly used. Applying antibiotics is optional since they can cause artifacts. Chloramphenicol is a commonly used drug to arrest elongating ribosomes in bacterial RIBO-seq. However, it does not prevent initiation, resulting...
Disclosures
The authors have nothing to disclose.
Acknowledgements
ALS would like to acknowledge the financial support of EMBO Installation Grants IG 3914, and POIR. 04.04.00-00-3E9C/17-00 carried out within the First TEAM programme of the Foundation for Polish Science co-financed by the European Union under the European Regional Development Fund.
Materials
| Name | Company | Catalog Number | Comments |
| 10X TBE (powder) | Invitrogen | AM9864 | |
| 2-Mercaptoethanol, 99%, pure | Acros Organics | 125472500 | |
| Adenosine 5'-Triphosphate (ATP) | New England Biolabs | P0756S | |
| Aluminium oxide calcinated pure p.a. | Chempur | 114560600 | |
| Calcium chloride dihydrate | Sigma-Aldrich | C3881-500G | |
| Chloramphenicol | MP Biomedicals | 190321 | |
| DNA Clean & Concentrator -5 | Zymo Research | D4004 | |
| Dnase I recombinant, Rnase-free | Roche | 4716728001 | |
| EDTA disodium salt | Fisher Scientific | E/P140/48 | |
| Ethyl Alcohol Absolut 99,8% Pure-P.A.-Basic | POCH Avantor Performance Materials Poland S.A | BA6480111 | |
| Filtration apparatus | VWR Collection | 511-0265 | all-glass filtration apparatus, with funnel, fritted base, cap, 47 mm Ø spring clamp and ground joint flask |
| Gel 40 (19:1) | Rotiphorese | 3030.1 | |
| Gel Loading Dye, Blue, 6X | New England Biolabs | E6138G | |
| Guanosine 5′-[β,γ-imido]triphosphate trisodium salt hydrate | Sigma-Aldrich | G0635-25MG | |
| labZAP | A&A Biotechnology | 040-500 | |
| Magnesium acetate tetrahydrate | Sigma-Aldrich | M5661-250G | |
| MCE membrane fiter | Alfatec Technology | M47MCE45GWS | pore size: 0.45um |
| MICROBExpress Bacterial mRNA Purification | Invitrogen | AM1905 | |
| Multiplex Small RNA Library Prep Set for Illumina | New England Biolabs | E7300S | |
| Nuclease-Free Water | Ambion | AM9937 | |
| Potassium Acetate Anhydrous Pure P.A. | POCH Avantor Performance Materials Poland S.A | 744330113 | |
| Quick-Load pBR322 DNA-MspI Digest | New England Biolabs | E7323A | |
| RNA Clean & Concentrator -25 | Zymo Research | R1018 | |
| Sodium acetate | Sigma-Aldrich | S2889-250G | |
| Sodium carbonate | Sigma-Aldrich | 223530-500G | |
| Sodium hydrogen carbonate pure p.a. | POCH Avantor Performance Materials Poland S.A | 810530115 | |
| SYBR Gold nucleic acid gel stain | Life Technologies | S11494 | |
| T4 Polynucleotide Kinase | New England Biolabs | M0201L | |
| T4 Polynucleotide Kinase Reaction Buffer | New England Biolabs | B0201S | |
| TBE-Urea Sample Buffer (2x) | Invitrogen | LC6876 | |
| Tris(hydroxymethyl)amino-methane, ultrapure, 99,9% | AlfaAesar | J65594 | |
| Triton X-100, 98% | Acros Organics | 327371000 | |
| Urea G.R. | lach:ner | 40096-AP0 |
References
- Ingolia, N. T., Ghaemmaghami, S., Newman, J. R., Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324 (5924), 218-223 (2009).
- Takanami, M., Yan, Y., Jukes, T. H. Studies on the site of ribosomal binding of f2 bacteriophage RNA. Journal of Molecular Biology. 12 (3), 761-773 (1965).
- Steitz, J. A. Polypeptide Chain Initiation: Nucleotide Sequences of the Three Ribosomal Binding Sites in Bacteriophage R17 RNA. Nature. 224 (5223), 957-964 (1969).
- Wolin, S. L., Walter, P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. The EMBO journal. 7 (11), 3559-3569 (1988).
- Argüello, R. J., et al. SunRiSE - measuring translation elongation at single-cell resolution by means of flow cytometry. Journal of Cell Science. 131 (10), 214346 (2018).
- Michel, A. M., Baranov, P. V. Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. Wiley Interdisciplinary Reviews: RNA. 4 (5), 473-490 (2013).
- Woolstenhulme, C. J., Guydosh, N. R., Green, R., Buskirk, A. R. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell reports. 11 (1), 13-21 (2015).
- McGlincy, N. J., Ingolia, N. T. Transcriptome-wide measurement of translation by ribosome profiling. Methods (San Diego, Calif). 126, 112-129 (2017).
- Gerashchenko, M. V., Gladyshev, V. N. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic acids research. 42 (17), 134 (2014).
- Ingolia, N. T., Lareau, L. F., Weissman, J. S. Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity of Mammalian Proteomes. Cell. 147 (4), 789-802 (2011).
- Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M., Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature protocols. 7 (8), 1534-1550 (2012).
- Hsu, P. Y., et al. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 113 (45), 7126-7135 (2016).
- Blanchard, S. C., Kim, H. D., Gonzalez, R. L., Puglisi, J. D., Chu, S. tRNA dynamics on the ribosome during translation. Proceedings of the National Academy of Sciences of the United States of America. 101 (35), 12893-12898 (2004).
- Becker, A. H., Oh, E., Weissman, J. S., Kramer, G., Bukau, B. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nature protocols. 8 (11), 2212-2239 (2013).
- Li, G. W., Oh, E., Weissman, J. S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 484 (7395), 538-541 (2012).
- King, H. A., Gerber, A. P. Translatome profiling: methods for genome-scale analysis of mRNA translation. Briefings in functional genomics. 15 (1), 22-31 (2014).
- Mohammad, F., Woolstenhulme, C. J., Green, R., Buskirk, A. R. Clarifying the Translational Pausing Landscape in Bacteria by Ribosome Profiling. Cell reports. 14 (4), 686-694 (2016).
- Reid, D. W., Shenolikar, S., Nicchitta, C. V. Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods (San Diego, Calif). 91, 69-74 (2015).
- Sanz, E., Yang, L., Su, T., Morris, D. R., McKnight, G. S., Amieux, P. S. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America. 106 (33), 13939-13944 (2009).
- Petrova, O. E., Garcia-Alcalde, F., Zampaloni, C., Sauer, K. Comparative evaluation of rRNA depletion procedures for the improved analysis of bacterialbiofilm and mixed pathogen culture transcriptomes. Scientific Reports. 7, 41114 (2017).
- Oh, E. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 147 (6), 1295-1308 (2011).
- Chew, G. L. Ribosome profiling reveals resemblance between long non-coding RNAs and 5' leaders of coding RNAs. Development. 140 (13), 2828-2834 (2013).
- Dunn, J. G., Foo, C. K., Belletier, N. G., Gavis, E. R., Weissman, J. S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2, 01179 (2013).
- Liu, M. J., et al. Translational landscape of photomorphogenic Arabidopsis. The Plant cell. 25 (10), 3699-3710 (2013).
- Rooijers, K., Loayza-Puch, F., Nijtmans, L. G., Agami, R. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nature Communications. 4, 2886 (2013).
- Danielle, L., et al. Fidelity of translation initiation is required for coordinated respiratory complex assembly. Science Advances. 5 (12), 2118 (2019).
- Zoschke, R., Watkins, K. P., Barkan, A. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. The Plant cell. 25 (6), 2265-2275 (2013).
- Chotewutmontri, P., Barkan, A. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS genetics. 12 (7), 1006106 (2016).
- Lee, S., Liu, B., Huang, S. X., Shen, B., Qian, S. B. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proceedings of the National Academy of Sciences of the United States of America. 109 (37), 2424-2432 (2012).
- Martinez, T. F., Chu, Q., Donaldson, C., Tan, D., Shokhirev, M. N., Saghatelian, A. Accurate annotation of human protein-coding small open reading frames. Nature chemical biology. 16 (4), 458-468 (2020).
- Simsek, D., et al. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell. 169 (6), 1051-1065 (2017).
- Fritsch, C., et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome research. 22 (11), 2208-2218 (2012).
- Lareau, L. F., Hite, D. H., Hogan, G. J., Brown, P. O. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife. 3, 01257 (2014).
- Meydan, S., et al. Retapamulin-Assisted Ribosome Profiling Reveals the Alternative Bacterial Proteome. Molecular Cell. 74 (3), 481-493 (2019).
- Wilson, D. N. The A-Z of bacterial translation inhibitors. Critical Reviews in Biochemistry and Molecular Biology. 44, 393-433 (2009).
- Nakahigashi, K. Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA research : an international journal for rapid publication of reports on genes and genomes. 23 (3), 193-201 (2016).
- Weaver, J., Mohammad, F., Buskirk, A. R., Storz, G. Identifying Small Proteins by Ribosome Profiling with Stalled Initiation Complexes. mBio. 10 (2), 02819 (2019).
- Nakahigashi, K. Effect of codon adaptation on codon-level and gene-level translation efficiency in vivo. BMC genomics. 15 (1), 1115 (2014).
- Li, G. W., Burkhardt, D., Gross, C., Weissman, J. S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 157 (3), 624-635 (2014).
- Michel, A. M., et al. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome research. 22 (11), 2219-2229 (2012).
- Guydosh, N. R., Green, R. Dom34 rescues ribosomes in 3' untranslated regions. Cell. 156 (5), 950-962 (2014).
- Young, D. J., Guydosh, N. R., Zhang, F., Hinnebusch, A. G., Green, R. Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3'UTRs In Vivo. Cell. 162 (4), 872-884 (2015).
- Ingolia, N. T. Ribosome profiling: new views of translation, from single codons to genome scale. Nature Reviews Genetics. 15 (3), 205-213 (2014).
- Dobin, A., et al. ultrafast universal RNA seq aligner. Bioinformatics. 29 (1), 15-21 (2013).
- . . Babraham Bioinformatics. Fast QC. , (2021).
- Gerashchenko, M. V., Gladyshev, V. N. Ribonuclease selection for ribosome profiling. Nucleic acids research. 45 (2), 6 (2017).
- Verbruggen, S., Menschaert, G. mQC: A post-mapping data exploration tool for ribosome profiling. Computer Methods and Programs in Biomedicine. 181, 104806 (2019).
- Cui, H., Hu, H., Zeng, J., Chen, T. DeepShape: estimating isoform-level ribosome abundance and distribution with Ribo-seq data. BMC bioinformatics. 20, 678 (2019).
- Choi, J. . RiboToolkit: an integrated platform for analysis and annotation of ribosome profiling data to decode RNA translation at codon resolution. , (2021).
- Choi, J. Dynamics of the context-specific translation arrest by chloramphenicol and linezolid. Nature chemical biology. 16 (3), 310-317 (2020).
- Tompson, J., O'Connor, M., Mills, J. A., Dahlberg, A. E. The protein synthesis inhibitors, oxazolidinones and chloramphenicol, cause extensive translational inaccuracy in vivo. Journal of Molecular Biology. 322 (2), 273-279 (2002).
- Brar, G. A., Weissman, J. S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nature reviews. Molecular cell biology. 16 (11), 651-664 (2015).
- Glaub, A., Huptas, C., Neuhaus, K., Ardern, Z. Recommendations for bacterial ribosome profiling experiments based on bioinformatic evaluation of published data. Journal of Biological Chemistry. 295, 8999-9011 (2020).
- Sorour, M. H., Hani, H. A., Shaalan, H. F., El-Sayed, M. M. H. Experimental screening of some chelating agents for calcium and magnesium removal from saline solutions. Desalination and Water Treatment. 57 (48-49), 22799-22808 (2015).
- Mohammad, F., Green, R., Buskirk, A. R. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. Elife. 8, 42591 (2019).
- O'Connor, P. B., Li, G. W., Weissman, J. S., Atkins, J. F., Baranov, P. V. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 29 (12), 1488-1491 (2013).
- Protocol for RNA Clean & Concentrator -25. Zymo Research Available from: https://files.zymoresearch.com/protocols/_r1017_r1018_rna_clean_concentrator-25.pdf (2021)
- Protocol for use with NEBNext Small RNA Library Prep Set for Illumina (E7300, E7580, E7560, E7330). New England Biolabs Available from: https://www.international.neb.com/protocols/2018/03/27/protocol-for-use-with-nebnext-small-rna-library-prep-set-for-illumina-e7300-e7580-e7560-e7330 (2021)
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved