A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Using Next Generation Sequencing to Identify Mutations Associated with Repair of a CAS9-induced Double Strand Break Near the CD4 Promoter
In This Article
Summary
Presented here is sgRNA/CAS9 endonuclease and next-generation sequencing protocol that can be used to identify the mutations associated with double strand break repair near the CD4 promoter.
Abstract
Double strand breaks (DSBs) in DNA are the most cytotoxic type of DNA damage. Because a myriad of insults can result in these lesions (e.g., replication stress, ionizing radiation, unrepaired UV damage), DSBs occur in most cells each day. In addition to cell death, unrepaired DSBs reduce genome integrity and the resulting mutations can drive tumorigenesis. These risks and the prevalence of DSBs motivate investigations into the mechanisms by which cells repair these lesions. Next generation sequencing can be paired with the induction of DSBs by ionizing radiation to provide a powerful tool to precisely define the mutations associated with DSB repair defects. However, this approach requires computationally challenging and cost prohibitive whole genome sequencing to detect the repair of the randomly occurring DSBs associated with ionizing radiation. Rare cutting endonucleases, such as I-Sce1, provide the ability to generate a single DSB, but their recognition sites must be inserted into the genome of interest. As a result, the site of repair is inherently artificial. Recent advances allow guide RNA (sgRNA) to direct a Cas9 endonuclease to any genome locus of interest. This could be applied to the study of DSB repair making next generation sequencing more cost effective by allowing it to be focused on the DNA flanking the Cas9-induced DSB. The goal of the manuscript is to demonstrate the feasibility of this approach by presenting a protocol that can define mutations that stem from the repair of a DSB upstream of the CD4 gene. The protocol can be adapted to determine changes in the mutagenic potential of DSB associated with exogenous factors, such as repair inhibitors, viral protein expression, mutations, and environmental exposures with relatively limited computation requirements. Once an organism's genome has been sequenced, this method can be theoretically employed at any genomic locus and in any cell culture model of that organism that can be transfected. Similar adaptations of the approach could allow comparisons of repair fidelity between different loci in the same genetic background.
Introduction
Maintaining genomic stability is critical for all living organisms. Accurate DNA replication and a robust DNA damage response (DDR) are necessary to faithfully propagate the genetic material1,2. DNA damages occur regularly in most cells2,3. When these damages are sensed, cell cycle progression is halted, and DNA repair mechanisms are activated. Double strand breaks in DNA or DSBs are the most toxic and mutagenic type of DNA damage3,4.
While several DDR signaling pathways can repair these lesions, the most thoroughly studied DSB repair pathways are homologous recombination (HR) and non-homologous end joining (NHEJ). HR is a largely error-free pathway that repairs a DSB using a sister chromatid as a homologous template. This tends to happen in the S phase and G2 phase of a cell cycle5,6,7. NHEJ is more error-prone, but it can happen throughout the cell cycle8,9. Various reporter assays have been developed to measure the efficiency of specific repair mechanisms10,11,12. These assays tend to rely on flow cytometry for a high throughput measurement of DSB repair pathway activity using GFP or mCherry as a readout11,13. While highly efficient, they rely on canonical repair occurring at an artificially introduced DSB.
There are a variety of other methods used to study DSB repair. Many of these rely on immunofluorescence (IF) microscopy1,14. IF microscopy detects discrete nuclear foci representative of repair complexes after DSBs are induced by exposure to genotoxic chemicals or ionizing radiation15,16. Tracking the formation and resolution of these foci provides an indication of repair initiation and completion, respectively14,17. However, these methods of DSB induction (i.e., chemicals or ionizing radiation) do not cause DSBs at defined locations in the genome. It is also functionally impossible to use them to consistently induce only a small number (e.g., 2-4) of DSBs. As a result, the most commonly used methods of inducing DSBs cause a multitude of lesions randomly distributed throughout the genome18. A small number of DSBs can be introduced by inserting the recognition site for a rare-cutting endonuclease and expressing the pertinent endonuclease, such as I-Sce119. Unfortunately, the required integration of a target site prevents the examination of DSB at endogenous genomic loci.
This manuscript describes a method to detect mutations associated with the repair of a DSB generated at a user-defined locus. We provide a representative example of the approach applied to assess the ability of a viral protein to increase the number of mutations associated with a DSB. Specifically, this manuscript describes the use of a single guide RNA (sgRNA) to direct a CAS9 endonuclease to induce a DSB at human CD4 open reading frame in human foreskin keratinocytes expressing vector control (HFK LXSN) and HFK that expresses the E6 protein of human papilloma virus type 8 (HFK 8E6). Targeted next-generation sequencing (NGS) of the region surrounding the break allows mutations associated with the repair of the lesion to be rigorously defined. These data demonstrate that the viral protein causes an approximately 20-fold increase in mutations during DSB repair. It also provides an unbiased characterization of the mutagenic consequences of DSBs at a single locus without the need for whole-genome sequencing. In principle, the protocol could be readily adapted to compare the relative risk of mutations between genome loci or cell lines.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Cell plating
- Grow HFK LXSN and HFK 8E6 cells in 10 cm plates in keratinocyte culture media (10 mL/plate) with human keratinocyte growth supplement (HKGS) and 1% penicillin/streptomycin. Grow cells to about 80% confluence at 37 °C in a jacket incubator with 5% CO2.
- Replace culture media with 3 mL of trypsin-EDTA (0.05%, ethylenediamine tetraacetic acid). Incubate at 37 °C for 3 min. Neutralize trypsin with equal volume of fetal bovine serum (FBS) supplemented media and transfer cells to a 15 mL centrifuge tube. Centrifuge at 300 x g for 5 min.
- Resuspend cells with 10 mL of keratinocyte culture media with HKGS. Determine the concentration of cells with a hemocytometer.
- Plate 4 x 105 cells/6-cm plates (seed two plates for HFK LXSN and two plates for HFK 8E6) in 4mL of keratinocyte culture media with HKGS and 1% penicillin and streptomycin. Grow at 37 °C in a jacket incubator with 5% CO2.
NOTE: Analysis of HFK cells was chosen for this protocol for two reasons. First, HFK is a difficult to transfect cell line. Thus, by demonstrating that the protocol works in this cell line, evidence is provided that it will likely work in more commonly used and more readily transfected cells. Secondly, previously published data demonstrate that a viral protein (8E6) hinders the repair of double strand breaks in DNA20,21,22. Thus, comparing HFK LXSN and HFK 8E6 allows us to demonstrate the ability of the assay to detect increases in mutations associated with a reduction in cellular repair capacity.
2. Transfection
- On the day of transfection (24 h after plating), replace media with 3 mL of antibiotic-free supplemented media. Incubate for 2 h at 37 °C in a jacket incubator.
- Transfect cells with appropriate lipid-based transfection reagents according to the manufacturer's instructions.
- Warm transfection reagents to room temperature and pipette gently before using.
- For each cell line (HFK LXSN and HFK 8E6), place appropriate amount of transfection buffer (as directed by the manufacturer) in a sterile 1.5 mL centrifuge tube (Tube 1). Include another tube with the same amount of transfection buffer (mock transfection or Tube 2).
- Add 2 µg of plasmid DNA expressing CAS9/sgRNA targeting human CD4 (px330-CD4, 5'- GGCGTATCTGTGTGAGGACT) to Tube 1 from step 2.2.2. Pipette gently to mix completely. Add equal volume of sterile water to Tube 2.
- Include a control plate with transfection reagents alone (no plasmid) for each cell line.
NOTE: The second plate serves as a negative control in the experiment, allowing the user to confirm that transfection with the CAS9/sgRNA are not responsible for any mutations. - Add appropriate amount of the transfection reagent (as directed by the manufacturer) to the tube with DNA mixture (Tube 1) from step 2.2.3 and the mock transfection (Tube 2) from step 2.2.2. Pipette gently to mix completely. Incubate at room temperature for 15-30 min to allow sufficient time for complexes to form.
- Add the transfection mixture drop-wise to the plate. Gently rock the culture for 1 min to evenly distribute the transfection mixture.
- Incubate for 48 h after the transfection to allow CAS9 expression.
- Harvest cells by trypsinization.
- Replace culture media with 1 mL of trypsin-EDTA (0.05%, ethylenediamine tetraacetic acid). Incubate at 37 °C for 3 min. Neutralize trypsin with equal volume of FBS supplemented media.
- For each plate of cells, transfer the cell suspension to two microcentrifuge tubes with equal aliquots. Centrifuge at 300 x g for 5 min.
- Resuspend the cell pellet from one tube in step 2.4.2 in 1 mL of phosphate buffered saline (PBS) for sequencing. Resuspend the other tube from step 2.4.2 with ice-cold PBS for immunoblot.
- Harvest the whole cell lysates for immunoblot.
- Centrifuge the tube at 300 x g for 5 min. Discard the supernatant.
- Add 100 µL radioimmunoprecipitation assay buffer (RIPA lysis buffer) mixed with 1 % protease inhibitor and 1% phosphatase inhibitor into the tube, mix thoroughly with a pipette and incubate for 10 min on ice.
NOTE: RIPA lysis buffer contains 10 mM Tris-HCl, pH 8.0; 1 mM EDTA; 0.5 mM EGTA; 1% Triton X-100; 0.1% Sodium Deoxycholate; 0.1% SDS; 140 mM NaCl, and deionized water. - Centrifuge lysates at 13,000 x g for 10 min. Collect supernatants for immunoblot.
3. Measuring CAS9 expression via immunoblot
- Determine the protein concentration with a bicinchoninic acid (BCA) assay according to the manufacturer's instructions.
- Run 20 µg of protein of each sample in the wells of a 3%-8% Tris-acetate gel for 150 min and semidry transfer (10 V for 30 min and then 25 V for 12 min) to polyvinylidene difluoride membrane.
- After blocking the membrane in 5% nonfat dry milk in PBS with 0.1% tween (PBST) for 1 h at room temperature, add anti-CAS9 (1:1000) and anti-GAPDH (1:1000) antibodies. Incubate at 4 °C overnight.
- After washing the membrane with PBST, incubate the membrane with secondary antibody in 5% nonfat dry milk in PBST for 1 h at room temperature.
- Image the blot and determine CAS9 level by densitometry23. See Figure 1 for a representative blot.
NOTE: Detecting phosphorylated H2AX (S139) foci formation by immunofluorescence microscopy can be used to validate CAS9 activity14. A low number of distinct foci (typically 1-4 foci) are expected depending on the cell cycle position, whether mutations in the CAS9 target site prevent further cutting, and how many copies of the CAS9 cutting site exist in the genome of interest. A representative image is shown in Figure 2.
4. Nucleic acid extraction and amplicon generation
- Extract DNA from cell samples from step 2.4.3 using a high-molecular weight DNA extraction kit, as specified by the manufacturer.
- Resuspend primers with indicated solvent according to the datasheet. Dilute with the same reagent to 20 µM and pool 20 µM primers into the indicated pools.
NOTE: Primer pool is listed in Supplemental Table 1. - Create a PCR Master mix using a long amplification Taq polymerase for each 20 µM primer pool as specified in Table 1.
- Add 21 µL of the mastermixes to separate PCR tubes.
- Add 4 µL of the target sample (100 ng/µL) from step 4.1 to PCR assay tubes containing mastermix and cap assay tubes. Ensure separate reactions for each primer pool.
- Vortex to mix PCR assay tubes and centrifuge (quick spin) to remove droplets from tube lids.
- Place the PCR tubes on a conventional thermal cycler machine.
- Program PCR machine as specified in Table 2.
- Run the program on a thermal cycler.
5. PCR clean-up
- Remove primers from PCR reactions using a bead-based PCR cleanup system.
- Remove clean-up beads from the refrigerator 30 min prior to use.
- Vortex beads well prior to use and ensure all beads are resuspended.
- Add 30 µL (1.2x) of resuspended beads to each well of a deep well 96-well plate.
- Add 25 µL of PCR reaction to wells containing beads.
- Place the plate on a plate shaker at 2000 rpm for 2 min.
- Allow the plate to remain at room temperature for 5 min following shaking.
- Place the deep well plate on a 96-well plate magnet and incubate for 2 min.
- Remove and discard the supernatant without the disturbing beads.
- While the plate remains on the magnet, add 180 µL of 80% ethanol and incubate for 30 s. Remove and discard the supernatant.
- Repeat step 5.1.9.
- Using a 10 µL pipette, remove and discard any remaining liquid from the wells.
- Allow beads to dry at room temperature for 10 min.
- Add 20 µL of nuclease free water to the wells containing beads and remove the plate from the magnet.
- Shake the plate at 2000 rpm for 2 min at room temperature.
- Incubate the plate at room temperature for 5 min.
- Place the plate on a magnet stand and incubate for 2 min at room temperature.
- Remove the supernatant into a second, labeled PCR plate. This contains the cleaned-up DNA.
- Measure the concentration of each reaction with a Fluorometer.
- Ensure that dsDNA Fluorometer reagents are at room temperature.
- Set up Fluorometric assay tubes plus two additional tubes for standards.
- Add 199 µL of 1x dsDNA working solution to all but two tubes. Add 190 µL of the working solution to the last two tubes.
- Add 10 µL of the two standards (included in the Table of Materials) to separate the assay tubes.
- Add 1 µL of each PCR reaction to Fluorometer mastermix tubes.
- Vortex tubes to mix and incubate at room temperature for 2 min.
- On the home screen of the fluorometer, select the button with the assay kitin use (1x dsDNA) then select Read Standards and Run Samples.
- Insert standard 1 tube, select the Read button and then repeat for standard 2.
- Following step 5.2.6, repeat for one sample, select a sample volume of 1 µL and the resulting concentration will be provided.
- Repeat step 5.2.7 for the remaining samples.
- Calculate the projected molarity of all reactions and pool equal concentrations of reactions from each individual sample separately (one final pool per sample) using the equation below.
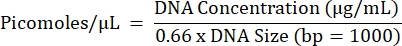
- Repeat steps 5.2.1 to 5.2.7 to obtain the final pool concentration.
- Check the amplicon pool on a capillary electrophoresis machine/agarose gel as specified by the manufacturer.
- Prepare capillary electrophoresis tubes for the appropriate number of samples. Add 7 µL of the DNA buffer as specified by the manufacturer.
- Add 4 µL of the amplicon pool to the tube containing DNA buffer.
- Place tubes on the electrophoresis machine and run the machine as specified by the manufacturer for dsDNA.
- View the electrophoresis gel pictures ensuring the bands localize to ~5 kb (size of amplicons).
- Calculate the projected molarity of all reactions and pool equal concentrations of reactions from each individual sample separately (one final pool per sample) using the equation below.
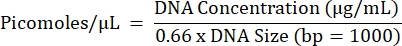
6. Library preparation
- Dilute sample pools from step 5.3.1 to 0.2 ng/µL for library preparation.
- Using a low-input library preparation kit compatible with short sequences (300bp) prepare libraries using unique index combinations for each sample pool created in step 5.3 following the manufacturer's instructions.
NOTE: Follow the manufacturer's instructions to select index sequences. All indexes amenable to the library prep kit will work for the samples. - Following library preparation, pool all samples according to the manufacturer's instructions.
NOTE: Before creating the library pool, calculate the number of reads necessary for 250x coverage of your target sequence and ensure that the selected sequencing cartridge can provide adequate coverage for each included sample. For 0.5Mb total, this will equate to 1M reads.
- Using a low-input library preparation kit compatible with short sequences (300bp) prepare libraries using unique index combinations for each sample pool created in step 5.3 following the manufacturer's instructions.
- Prepare the library pool for sequencing.
- Thaw and prepare a 300-cycle cartridge and sequencing reagents.
- Denature and dilute the sequencing pool created in step 5.1.1 according to the sequencer's manufacturer's instructions.
- Add denatured and diluted library pool to sequencing reagents and Run the sequencing machine as specified by the manufacturer.
NOTE: See attached Table 3 for trouble shootings.
7. Data analysis
NOTE: All data steps are performed in the genomic data analysis software. Parentheses indicate user input. Greater than sign indicates the order of mouse clicks for any given step (e.g., 1st mouse click>2nd mouse click)
- Import the reads by clicking on Open Software > Import > Illumina > Select Files > Next > Select location to Save > Finish. The reads will now appear in the software.
- Trim and filter the reads.
- In the deep sequence data analysis software, trim the raw reads default parameters.
- Highlight the reads and click Toolbox > Prepare Sequencing Data > Trim Reads > Next > Next > Next > Next > Save > Next > (Select location to save) > Finish
- Map the trimmed reads to reference.
- Map trimmed reads to the reference sequence used in step 4.1 using a match score of 2, mismatch cost of 3 and insertion/deletion costs of 2. Ensure that the length fraction is above 0.7 and similarity fraction is at or above 0.8.
- Highlight the trimmed read file and click Toolbox > Resequencing Analysis > Map Reads to Reference > Next > (Select reference sequence) > Next > (Ensure parameters are indicated as above) > Next > Save > (Select location to save) > Finish.
- Extract variants and indels.
- Using an appropriate indel caller, extract indels using a p-value threshold of 0.005 or lower and a maximum number of mismatches of 3.
- Highlight mapped read file and click Toolbox > Resequencing Analysis > Variant detection > Indels and Structural Variants > Next > (Ensure required significance is input > Next > Save > (Select location to save) > Finish.
- Using an appropriate variant caller, call variants from the read mapping using a significance of 5%.
- Highlight mapped read file and click Toolbox > Resequencing Analysis > Variant Detection > Low frequency variant detection > Next > (Ensure required significance is input > Next > Next > Save > (Select location to save) > Finish.
NOTE: Ensure to account for the ploidy of the host genome in the indel and variant callers. Do not extract mutants below 5%. This threshold accounts for PCR and sequencing errors associated with the assay. Normalization (based on immunoblot detection of CAS9) should be done by adjusting sequencing coverage. For example, if sample A has twice the transfection efficiency of sample B, then 50% of the reads from sample A should be used for analysis. This should be done by random sampling and not reduce the coverage for any sample below 100x.
Access restricted. Please log in or start a trial to view this content.
Results
Three representative results are presented for this protocol. Figure 1 is an immunoblot confirming expression of CAS9 in HFK control (LXSN) and HFK expressing beta-HPV 8E6 (8E6). 48 h after transfection, whole cell lysates were harvested and subsequently probed with an anti-CAS9 antibody (or GAPDH as a loading control). The result shows that HFK LXSN and HFK 8E6 are expressing similar amount of CAS9 indicating that transfection efficiency is similar between t...
Access restricted. Please log in or start a trial to view this content.
Discussion
In addition to the depth of information provided, there are several advantages to this method. First, DSB repair, in theory, can be assessed at any genomic loci without modifying the genome of the cell of interest. Second, access to NGS analysis of repair is increased by the reduced cost and computational effort afforded by making and analyzing a single DSB targeted to a defined area. Finally, with the genomes of additional organisms routinely becoming available and multiple publications demonstrating successful transfec...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Research reported in this manuscript was supported by the National Institute of General Medical Sciences of the National Institutes of Health (P20GM130448) (NAW and RP); National Cancer Institute of the National Institutes of Health (NCI R15 CA242057 01A1); Johnson Cancer Research Center in Kansas State University; and the U.S. Department of Defense (CMDRP PRCRP CA160224 (NAW)). We appreciate KSU-CVM Confocal Core and Joel Sanneman for our immunofluorescence microscopy. The content is solely the responsibility of the authors and does not necessarily represent the official views of these funding agencies.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 6 Well Tissue Culture Plate | Celltreat | 229106 | Cell culture plate |
| BCA Kit | VWR | 89167-794 | BCA assay kit |
| Centrifuge 5910 R | Eppendorf | 2231000772 | Tabletop Centrifuge |
| CLC Genomic Workbench | Qiagen | 832001 | deep sequence data analysis software/indel caller/variant caller |
| Digital Microplate Genie pulse | Scientific industries | SI-400A | Plate shaker |
| DYKDDDDK Tag Monoclonal Antibody (FG4R) | ThermoFisher Scientific | MA191878 | Anti-FLAG antibody |
| Epilife CF Kit | ThermoFisher Scientific | MEPICF500 | Cell cultrue media and supplements |
| Fetal Bovine Serum (FBS) | VWR | 89510-194 | Cell culture supplement |
| Goat anti-Rabbit IgG | ThermoFisher Scientific | A-11012 | Secondary antibody |
| HighPrep PCR Clean-up system | MagBio | AC-60005 | Bead-based PCR cleanup kit |
| KAPA HiFi HotStart ReadyMix PCR Kit | KAPA Biosystems | KK2600 | PCR mastermix/PCR assay |
| MagAttract HMW DNA kit | Qiagen | 67563 | High Molecular Weight DNA extraction kit |
| Magnetic Stand-96 | Thermo Fisher Scientific | AM10027 | 96-Well Magnetic Rack |
| MiniAmp Thermal Cycler | Applied Biosystems | A37834 | Thermal Cycler |
| Miseq | Illumina | SY-410-1003 | Sequencer |
| Miseq v2 300 cycle reagent kit | Illumina | MS-102-2002 | 300-cycle cartridge/sequencing reagents |
| Nextera XT DNA Library Prep kit | Illumina | FC-131-1024 | Library preparation kit |
| Nextera XT Kit v2 Set A | Illumina | 20027215 | Indexes |
| Nunc 96-well polypropylene DeepWell Stroage plates | Thermo Fisher Scientific | 260251 | deep well 96-well plates |
| Penicillin-Streptomycin Solution (100X) | Calsson Labs | PSL02-6X100ML | Antibiotics for cell culture |
| Phosphate Buffered Saline (PBS) | Bio Basic | PD8117 | PBS |
| px330-CD4 | Addgen | 136938 | SgRNA/CAS9 plasmids targeting 5’- GGCGTATCTGTGTGAGGACT |
| QIAxcel Advanced System | Qiagen | 9001941 | capillary electrophersis machine |
| QIAxcel DNA screening kit | Qiagen | 929004 | DNA buffer/ capillary electrophersis tubes |
| Qubit 1x ds HS Assay Kit | ThermoFisher Scientific | Q23851 | Fluorometer reagents/1x dsDNA solution |
| Qubit 4 Fluorometer | ThermoFisher Scientific | Q33238 | Fluorometer |
| Qubit Assay Tubes | Thermo Fisher Scientific | Q32856 | Fluorometer assay tubes |
| RIPA Lysis Buffer | VWR | VWRVN653-100ML | Lysis buffer for protein extraction |
| Trypsin-EDTA (0.05%), phenol red | ThermoFisher Scientific | 25300054 | Trypsin |
| Vortex-Genie 2 | Scientific industries | SI-0236 | Vortex |
| Xfect Transfection Reagent | Takara Bio | 631318 | Transfection reagent |
| genomic data analysis software | QIAGEN | CLC Workbench v21.0. | Data analysis software |
References
- Vítor, A. C., Huertas, P., Legube, G., de Almeida, S. F. Studying DNA double-strand break repair: an ever-growing toolbox. Frontiers in Molecular Biosciences. 7, (2020).
- Giglia-Mari, G., Zotter, A., Vermeulen, W. DNA damage response. Cold Spring Harbor Perspectives in Biology. 3 (1), 000745(2011).
- Khanna, K. K., Jackson, S. P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nature Genetics. 27 (3), 247-254 (2001).
- vanden Berg, J. G., et al. A limited number of double-strand DNA breaks is sufficient to delay cell cycle progression. Nucleic Acids Research. 46 (19), 10132-10144 (2018).
- Chang, H. H. Y., Pannunzio, N. R., Adachi, N., Lieber, M. R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nature Reviews Molecular Cell Biology. 18 (8), 495-506 (2017).
- Daley, J. M., Sung, P. 53B. P. 1 BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Molecular and Cellular Biology. 34 (8), 1380-1388 (2014).
- Godin, S. K., Sullivan, M. R., Bernstein, K. A. Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication. Biochemistry and Cell Biology = Biochimie et Biologie Cellulaire. 94 (5), 407-418 (2016).
- Jette, N., Lees-Miller, S. P. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Progress in Biophysics and Molecular Biology. 117 (0), 194-205 (2015).
- Weterings, E., van Gent, D. C. The mechanism of non-homologous end-joining: A synopsis of synapsis. DNA Repair. 3 (11), 1425-1435 (2004).
- Bhargava, R., Lopezcolorado, F. W., Tsai, L. J., Stark, J. M. The canonical non-homologous end joining factor XLF promotes chromosomal deletion rearrangements in human cells. Journal of Biological Chemistry. 295 (1), 125-137 (2020).
- Gunn, A., Bennardo, N., Cheng, A., Stark, J. M. Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. Journal of Biological Chemistry. 286 (49), 42470-42482 (2011).
- Simsek, D., Jasin, M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nature Structural & Molecular Biology. 17 (4), 410-416 (2010).
- Certo, M. T., et al. Tracking genome engineering outcome at individual DNA breakpoints. Nature Methods. 8 (8), 671-676 (2011).
- Murthy, V., et al. Characterizing DNA repair processes at transient and long-lasting double-strand DNA breaks by immunofluorescence microscopy. JoVE Journal of Visualized Experiments. (136), e57653(2018).
- Wang, J. L., et al. Dissection of DNA double-strand-break repair using novel single-molecule forceps. Nature Structural & Molecular Biology. , (2018).
- Azzam, E. I., Jay-Gerin, J. -P., Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Letters. 327 (0), 48-60 (2012).
- Kuo, L. J., Yang, L. -X. γ-H2AX - A novel biomarker for DNA double-strand breaks. In Vivo. 5, (2008).
- Sanders, J. T., et al. Radiation-induced DNA damage and repair effects on 3D genome organization. Nature Communications. 11 (1), 6178(2020).
- Bellaiche, Y., Mogila, V., Perrimon, N. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics. 152 (3), 1037-1044 (1999).
- Wallace, N. A., Robinson, K., Howie, H. L., Galloway, D. A. β-HPV 5 and 8 E6 disrupt homology dependent double strand break repair by attenuating BRCA1 and BRCA2 expression and foci formation. PLOS Pathogens. 11 (3), 1004687(2015).
- Hu, C., Bugbee, T., Gamez, M., Wallace, N. A. Beta human papillomavirus 8E6 attenuates non-homologous end joining by hindering DNA-PKcs activity. Cancers. 12 (9), 2356(2020).
- Hu, C., Bugbee, T., Dacus, D., Palinski, R., Wallace, N. A. Beta human papillomavirus 8 E6 allows colocalization of non-homologous end joining and homologous recombination repair factors. PLOS Pathogens. 18 (3), 1010275(2022).
- Butler, T. A. J., Paul, J. W., Chan, E. -C., Smith, R., Tolosa, J. M. Misleading westerns: Common quantification mistakes in western blot densitometry and proposed corrective measures. BioMed Research International. 2019, 5214821(2019).
- Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S., Bonner, W. M. DNA double-stranded breaks induce histone H2AX Phosphorylation on serine 139. Journal of Biological Chemistry. 273 (10), 5858-5868 (1998).
- Taning, C. N. T., Van Eynde, B., Yu, N., Ma, S., Smagghe, G. CRISPR/Cas9 in insects: Applications, best practices and biosafety concerns. Journal of Insect Physiology. 98, 245-257 (2017).
- Ghezraoui, H., et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Molecular Cell. 55 (6), 829-842 (2014).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved