Method Article
Analytical Determination of Mitochondrial Function of Excised Solid Tumor Homogenates
In This Article
Summary
We developed a practical protocol and analytical approach to evaluate mitochondrial oxidative phosphorylation and electron transfer capacity in fresh tumor homogenates. This protocol can be easily adapted to survey various mitochondrial functions that contribute to cancer initiation, progression, and treatment response.
Abstract
Mitochondria are essential to the onset and progression of cancer through energy production, reactive oxygen species regulation, and macromolecule synthesis. Genetic and functional adaptations of mitochondria to the tumor environment drive proliferative and metastatic potential. The advent of DNA and RNA sequencing removed critical barriers to the evaluation of genetic mediators of tumorigenesis. However, to date, methodological approaches to evaluate tumor mitochondrial function remain elusive and require technical proficiency limiting the feasibility, ultimately diminishing diagnostic and prognostic value in both experimental and clinical settings. Here, we outline a simple and rapid method to quantify rates of oxidative phosphorylation (OXPHOS) and electron transfer (ET) capacity in freshly excised solid tumor homogenates using high-resolution respirometry. The protocol can be reproducibly applied across species and tumor types as well as adapted to evaluate a diversity of mitochondrial ET pathways. Using this protocol, we demonstrate that mice bearing a luminal B mammary cancer exhibit defective nicotinamide adenine dinucleotide-linked respiration and reliance on succinate to generate adenosine triphosphate via OXPHOS.
Introduction
All cells are intimately linked by their need to produce and consume adenosine triphosphate (ATP), the molecular energy currency. As cellular mutations give rise to the formation of tumors, mitochondria ensure survival through diversification of energy production that is often phenotypically distinguishable from non-cancerous tissue1,2,3. As such, there is a critical need for rapid and deep profiling of mitochondrial respiratory function in order to facilitate the classification of tumor type, cancer initiation, progression, and treatment response.
Respiratory functions of excised tissue specimens cannot be evaluated intact as the primary substrates for OXPHOS are not cell-permeable. To overcome this limitation, mitochondria can be prepared either by isolation, chemical permeabilization, or mechanical homogenization. Mitochondrial isolation is long considered to be a gold standard for the evaluation of respiratory function. However, it requires large amounts of tissue, is time-consuming, and is low-yielding with possible selection bias for certain fractions of mitochondria4. Permeabilization consists of mechanical separation and exposure of tissue sections or fiber bundles to a mild detergent that selectively degrades the plasma membrane5. Permeabilization is frequently employed in striated tissues such as skeletal and cardiac muscle as individual fiber bundles can be teased apart. Compared to isolation, permeabilization yields more mitochondria in their native cellular environment and physical form5. Permeabilization has been successfully applied in other tissues such as tumor6,7 and placenta8; however, reproducibility of permeabilized fiber preparations can be difficult due to consistency of dissection and oxygen requirements to overcome diffusion limitations9. Additionally, permeabilized fibers may be unsuitable in certain tumor types that are densely cellular and highly fibrotic. Tissue homogenates are generated through mechanical disruption of the plasma membrane and are similar to permeabilized fibers in terms of mitochondrial yield and integrity10. Tissue homogenates also minimize the limitations of oxygen diffusion and can be readily employed across tissue types through optimization of mechanical force11,12.
Here, we outline a simple and rapid method to quantify rates of oxidative phosphorylation (OXPHOS) and electron transfer (ET) capacity in freshly excised solid tumor homogenates. The protocol is optimally designed to evaluate fresh tissue using the Oxygraph-2k (O2k) high-resolution respirometer, which requires prior knowledge of instrumental setup and calibration but can be similarly adapted using any Clark-type electrode, Seahorse analyzer, or plate reader. The protocol can be reproducibly applied across species and tumor types as well as adapted to evaluate a diversity of mitochondrial ET pathways.
Protocol
All experiments and procedures involving animals were approved by the Pennington Biomedical Research Center Institutional Animal Care and Use Committee.
1. Reagent preparation.
- Prepare EO771 cell growth media with 10 mM HEPES, 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (100 U/mL), and 0.2% amphotericin B.
- Prepare 1 L of biopsy preservation (BIOPS) solution in a 1000 mL glass beaker.
- Add 3.180 g of Na2ATP (5.77 mM final concentration).
- Add 1.334 g of MgCL2·6H2O (6.56 mM final concentration).
- Add 2.502 g of Taurine (20 mM final concentration).
- Add 3.827 g of Na2Phosphocreatine (15 mM final concentration).
- Add 1.362 g of Imidazole (20 mM final concentration).
- Add 0.077 g of Dithiothreitol (.5 mM final concentration).
- Add 9.76 g of MES hydrate (50 mM final concentration).
- Add 800 mL of H2O and mix the constituents using a magnetic stirrer at 30 °C.
- Add 72.3 mL of 100 mM K2EGTA (7.23 mM final concentration).
- Dissolve 7.608 g of EGTA and 2.3 g of KOH in 100 mL of H2O.
- Adjust pH to 7.0 with 5 M KOH and bring the volume up to 200 mL with H2O.
- Add 27.7 mL of 100 mM CaK2EGTA (2.77 mM final concentration).
- Dissolve 7.608 g of EGTA in 200 mL of H2O and heat to 80 °C (100 mM final concentration).
- Dissolve 2.002 g of CaCO3 in 200 mL of hot 100 mM EGTA solution.
- While continuously stirring, add 2.3 g of KOH and adjust the pH to 7.0.
- Adjust the pH to 6.75 at 23 °C (pH 7.1 at 0 °C) with 5 M KOH. Bring the volume up to 980 mL with H2O, and mix the solution. Check the pH once more, adjust if necessary, and bring the final volume up to 1000 mL with water.
- Aliquot BIOPS into conical tubes (15 mL or 50 mL) and store at -20 °C until use. Only thaw once, just before use.
- Prepare 1 L of mitochondrial respiration medium (MiR05) in a 1000 mL glass beaker.
- Add 0.190 g of EGTA (0.5 mM final concentration).
- Add 0.610 g of MgCL2·6H2O (3 mM final concentration).
- Add 2.502 g of Taurine (20 mM final concentration).
- Add 1.361 g of KH2PO4 (10 mM final concentration).
- Add 4.77 g of HEPES (20 mM final concentration).
- Add 37.65 g of D-Sucrose (110 mM final concentration).
- Add 800 mL of H2O and mix the constituents using a magnetic stirrer at 30 °C.
- Add 120 mL of 0.5 M Lactobionic acid (60 mM final concentration).
- Dissolve 35.83 g of Lactobionic acid in 100 mL of H2O.
- Adjust pH to 7.0 with 5 M KOH and bring volume up to 200 mL with H2O.
- Mix the solution and adjust the pH to 7.1 with 5 M KOH.
- Weigh 1 g of BSA, essentially fatty acid-free (1 g/L final concentration), in a 50 mL conical tube. Add 40 mL of the pH 7.1 solution from step 9 to tube, invert gently to mix, and avoid foaming. Transfer the dissolved BSA into the remaining pH 7.1 solution from step 9 and stir gently and continuously. Check pH once more, adjust if necessary, and bring the final volume up to 1000 mL with H2O.
- Aliquot the MiR05 medium into 50 mL conical tubes and store at -20 °C until use. Only thaw once, just before use.
- Prepare substrates, uncouplers, and inhibitors.
- Prepare 0.8 M Malate: Dissolve 536.4 mg of L-Malic acid in 4 mL of H2O. Neutralize with 5 M KOH to pH 7 and bring the volume up to 5 mL with H2O. Divide into aliquots and then store at -20 °C.
- Prepare 1 M Pyruvate: Dissolve 550 mg of Pyruvic acid sodium salt in 4 mL of H2O. Neutralize with 5 M KOH to pH 7 and bring the volume up to 5 mL with H2O. Divide into aliquots and then store at -20 °C.
- Prepare 0.5 M ADP (adenosine 5′-diphosphate): Dissolve 1.068 g of ADP sodium salt in 4 mL of H2O. Neutralize with 5 M KOH to pH 7 and bring the volume up to 5 mL with H2O. Divide into aliquots and then store at -20 °C.
- Prepare 2 M Glutamate: Dissolve 3.7426 g of L-Glutamic acid monohydrate in 8 mL of H2O. Neutralize with 5 M KOH to pH 7 and bring the volume up to 10 mL with H2O. Divide into aliquots and then store at -20 °C.
- Prepare 4 mM Cytochrome c: Dissolve 50 mg of Cytochrome c in 1 mL of H2O. Divide into aliquots and then store at -20 °C.
- Prepare 1 M Succinate: Dissolve 2.701 g of Succinate disodium salt hexahydrate in 8 mL of H2O. Neutralize with 1 N HCl to pH 7 and bring the volume up to 10 mL with H2O. Divide into aliquots and then store at -20 °C.
- Prepare 1 mM FCCP (carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone): Dissolve 2.54 mg of FCCP in 10 mL of pure ethanol. Divide into aliquots and then store at -20 °C.
- Prepare 150 µM Rotenone: Dissolve 3.94 mg of Rotenone in 10 mL of pure ethanol to prepare 1 mM stock, vortex until fully dissolved. Dilute 225 µL of 1 mM Rotenone stock concentration with 1.275 mL of pure ethanol to make 1.5 mL of 150 µM Rotenone. Protect from light, divide into aliquots, and then store at -20 °C.
- Prepare 125 µM Antimycin A: Dissolve 11 mg of Antimycin A in 4 mL of pure ethanol to prepare 5 mM stock concentration of Antimycin A. Dilute 25 µL of 5 mM Antimycin A stock concentration with 975 µL of pure ethanol to make 1 mL of 0.125 mM Antimycin A. Divide into aliquots and then store at -20 °C.
- Prepare 0.8 M Ascorbate: Dissolve 1.584 g of Ascorbate sodium salt in 8 mL of H2O. Adjust pH with ascorbic acid to pH 6 and bring the volume up to 10 mL with H2O. Protect from light, divide into aliquots, and then store at -20 °C.
- Prepare 0.2 M TMPD (tetramethyl-p-phenylenediamine): Dissolve 32.85 mg of TMPD in 987.5 µL of DMSO. Add 12.5 µL of 0.8 M ascorbate (10 mM final concentration of ascorbate). Protect from light, divide into aliquots, and then store at -20 °C.
- Prepare 4 M Sodium azide: Dissolve 2.6 g of Sodium Azide in 10 mL of H2O. Divide into aliquots and store at -20 °C.
2. Tumor growth
- Grow EO771 cells in RPMI 1640 growth media and maintain the cells in a 37 °C humidified incubator with 5% CO2.
- Single-house four-week old female C57BL/6J mice, maintain them at 21-22 °C on a 12 h light: dark cycle. Provide the mice ad libitum access to food and water.
- Once the mice reach 10 weeks of age, prepare the cells and the mice for cancer cell implantation.
- Trypsinize the cells, deactivate trypsin with growth media, and centrifuge cells at 500 x g for 5 min at room temperature. Aspirate the supernatant and resuspend and combine the cell pellets (as needed) in media. Count viable cells using trypan blue and prepare a cell dilution of 1 x 106 cells in a total volume of 60 µL with a 1:1:1 Media/ Basement Membrane Matrix/PBS solution. Once the basement membrane matrix is added, mix well and keep the cell suspension on ice. Fill syringes with 60 µL of cell suspension and place them on ice. Work efficiently and inject cells into mice within 1.5 h of preparation.
- Anesthetize mice by isoflurane inhalation (3%-5% for induction and 1%-3% for maintenance). Shave between the right 4th and 5th inguinal mammary glands. Orthotopically inject the anesthetized mice with the EO771 cell suspension.
- Use electronic calipers to monitor and measure the tumor growth twice a week for 4 weeks. At necropsy, excise, weigh, and then place the tumor (or a minimum of 60 mg tumor section) immediately into 10 mL of ice-cold BIOPS. Keep the tube on wet ice.
3. Instrument setup and calibration
- Warm the MiR05 medium in a 37 °C water bath.
- Turn on the Oroboros O2k system, open DATLAB software, enter or select User. Click on Connect to O2k. Check the O2k configuration and ensure that the correct instrument is labeled for Power-O2k and each chamber corresponds to the correct Oxygen Sensor; click OK. Once the O2k control window opens, under the Systems tab, set the Block Temperature to 37 °C, the Stirrer speed to 750 rpm for both chambers, ND the Data Recording interval to 2.0 s. Check both Stirrer Power and Illumination in Chamber boxes. In the Oxygen, O2 tab, set the Gain for Sensor to 1 V/µA (the gain may need to be adjusted for older instrument models), the Polarization Voltage to 800 mV, and then click on Connect to O2k. Once a new experiment file opens, name the file and click on Save. Once the experiment is active, a protocol selection window will open; click on Cancel to run a custom SUIT (Substrate uncoupler inhibitor titration).
- Following protocol selection, a sample window will open to fill out the experimental code, sample type, cohort, sample code, sample number, and subsample number as applicable. Assign the unit to mg and enter the tumor homogenate concentration per mL (amount per chamber will auto-populate). Ensure that the indicated Medium is MiR05 and the Chamber Volume is 2.00 mL. Add comments as needed in the bottom box and click on OK.
- Remove the stoppers and aspirate the 70% ethanol from the chambers. Never aspirate close to the membrane that is exposed on the inside of the chamber. Rinse four times with pure water and fill the chamber with 2.25 mL of MiR05.
- Sensor test: Click on F9 to switch off the stirrers for 30 s. Once the stirrer bar switches back on, ensure that the O2 slope rapidly increases monoexponentially in each chamber. If the chamber fails the sensor test, check the electrical connections and clean if salts have accumulated; repeat the test. If the sensor continues to fail the test, remove the polarographic oxygen sensor (POS) connector to inspect the membrane. If visible damage to the membrane, heavy oxidation, or significant bubble accumulation is observed, halt the experimental procedures, and proceed to instrument servicing.
- Oxygen calibration: Perform oxygen calibration to obtain accurate respiration measurements.
- With a twisting motion, Insert the stoppers slowly to their volume-calibrated position. Siphon off the excess medium ejected through the injection capillary that collects in the well of the stopper. With a twisting motion, lift the stoppers just enough to tightly fit the stopper-spacer tool leaving a gas volume above the liquid phase for the final air equilibration (30-45 min.).
- Use the steady-state achieved over this period to calibrate the oxygen sensor to obtain accurate measurements of respiration. The O2 slope neg. (negative slope of oxygen) is 0 ± 2 pmol/s·mL. If the value is higher than 2 pmol/s·mL, clean the chamber and replenish it with freshly thawed MiR05. If the slope is unstable, proceed to instrument servicing.
- After achieving a steady state, select the O2 concentration curve and highlight the steady region of the curve by holding down the shift key and left-click mouse button. Once the region is selected, click on F5, select the mark for air calibration with the drop-down arrow and click on Calibrate and copy to clipboard.
- Instrumental background calibration (optional)
- Following air calibration, ensure that there is no gas volume above the liquid phase for flux background equilibration (~15 min) and close the chambers fully.
NOTE: The steady-state rate achieved over this period represents instrumental background and can be subtracted from the resulting data for enhanced analytical precision. The O2 slope neg. will initially increase slightly and plateau between ± 2 to 4 pmol/s·mL. - If the value is high (>6 pmol/s·mL), there is potential biological contamination. In this case, clean the chamber and replenish it with freshly thawed MiR05. Once a steady-state is achieved, select the O2 slope neg. curve and highlight the steady region of the curve by holding down the Shift key and left-click mouse button.
- After selecting the region, click on F5, select the Baseline Correction Box and the mark for Baseline with the drop-down arrow, and click on OK.
- Following air calibration, ensure that there is no gas volume above the liquid phase for flux background equilibration (~15 min) and close the chambers fully.
4. Tumor homogenate preparation
- Place a glass homogenizer containing 1 mL of MiR05 and a tightly fitting glass pestle on wet ice.
- At the time of the study, place the tissue in 1 mL of BIOPS in an ice-cold Petri dish.
- Clean and dissect the tissue to maximize soluble material, avoid necrotic regions, and remove marginal tumor tissue. Using a dissection microscope, scalpel, and surgical tweezers, remove any hair, necrotic tissue, peripheral connective and vascular tissue, and adjacent fat, as applicable. Take care to keep the tumor in ice-cold BIOPS during the dissection.
- Cut the tumor into small pieces (~5-10 mg each) and place the remaining tumor pieces back into 10 mL of BIOPS kept on ice. Use this tissue later for additional preparations if needed.
NOTE: Perform the following steps quickly to minimize the amount of time the sample is not submerged fully in BIOPS. Once the sample is placed in MiR05, time is of the essence. Carefully move the prepared homogenate into the calibrated chamber as soon as possible. - Blot the tissue sections carefully on a filter paper, place them on a small plastic tared weigh boat, and record the initial wet weight. Place the unused pieces back into the 10 mL conical tube of BIOPS for continued preservation.
- Submerge the tissue sections into an ice-cold homogenizer containing MiR05 and record the remaining weight on the weigh boat, if applicable.
- Using a glass pestle (clearance range 0.09-0.16 mm), gently disrupt the tumor tissue by completing 5-7 down-and-up strokes. For each stroke, rotate the pestle in a clock-wise-counter-clockwise motion 3 times while pushing the pestle down and 3 additional times while pulling the pestle back up. Be sure to allow the tissue to settle to the bottom of the homogenizer in-between strokes but avoid bringing the pestle fully above the fluid volume to prevent foaming.
NOTE: The resulting homogenate should appear cloudy with minimal solid tissue remnants. - Pour the homogenate into a 15 mL conical tube and place it on ice.
- Pipette 1-3 mL of fresh MiR05 over the pestle and into the homogenizer to wash any remaining tissue homogenate. Pour the MiR05 wash into the conical tube containing the homogenate. Repeat the pestle and homogenizer wash 2-3 times to ensure complete transfer of the homogenate. Keep in mind the target concentration and required volume to not over dilute the sample during the washing steps.
- To accurately calculate the tissue homogenate concentration, carefully inspect the homogenizer and the homogenate for remaining non-homogenized material (i.e., connective tissue).
- To remove the non-homogenized material from the homogenizer that cannot be reached with tweezers, add MiR05 to the homogenizer, aspirate the volume (including the tissue) with a pipette and move the contents to a Petri dish.
- To remove the non-homogenized material from the homogenate (settled in large pieces at the bottom of the conical tube), aspirate the large pieces with a pipette and place them on the tissue cap of the conical tube.
- Remove the tissue from the petri dish or conical tube cap with tweezers and blot it on filter paper. Add the remaining homogenate from the conical tube cap back into the conical tube, cap it, and then invert to mix.
- Re-inspect the homogenizers and homogenate for any additional non-homogenized material, and repeat steps 4.10.1-4.10.3 to remove the material as needed.
- Weigh and record the mass of the non-homogenized material recovered from the homogenizer. Inspect the homogenate preparation for any grossly intact tissue transferred. Remove the non-homogenized pieces and weigh as necessary.
- Subtract the tissue weight recovered from the weigh boat, homogenizer, and homogenate (if necessary) from the initial wet weight to calculate the final sample weight.
- Using the final sample weight, add additional MiR05 to bring homogenate to the desired concentration (see step 5 for details regarding optimization experiments).
- Once the homogenate is weighed and prepared, proceed to assay as soon as possible. Keep the sample stored on wet ice until it is transferred to the instrument.
5. Substrate, uncoupler, inhibitor titration protocol (SUIT)
- Once the instrument is calibrated, and the sample is prepared, remove the stoppers with a twisting motion and aspirate the MiR05 from the chambers (avoiding the membrane exposed inside the chamber). Mix the homogenate well and add 2.25 mL of the homogenate to the chamber. If adding one homogenate to multiple chambers, pipette 1 mL at a time into each chamber while mixing the homogenate to ensure equal tissue distribution. Click on F4 to name and time stamp the event and then click on OK.
- Fill a 50 mL syringe with oxygen from an oxygen tank with a regulator and gas tubing using a blunt 18 G needle. Hyperoxygenate the chambers to ~500 µM oxygen. For this, inject oxygen directly into the chambers. Loosely insert the stoppers and wait to close until the oxygen reaches ~480 µM. With a twisting motion, slowly close the chamber and allow the respiration to equilibrate (~15-20 min). Fill the stopper central capillary with MiR05 if necessary.
- Analytical determination of the OXPHOS and ET (electron transfer state) capacity of N-linked and NS-linked and CIV (Complex IV) activity: Use dedicated microsyringes to inject substrates, uncouplers, and inhibitors into fully closed chambers. Click on F4 with each injection to name and time stamp the events for each chamber in real-time. Throughout the study, select F6 to adjust the O2 concentration and O2 slope neg. scales as needed. After each injection, wash the syringes 3 times in water (for water-soluble compounds) or 70% ethanol (for compounds dissolved in ethanol or DMSO).
NOTE: N-linked: O2 flux supported by defined NADH-generating substrate combinations, NS-linked: O2 flux supported by the convergence of defined NADH-generate substrate combinations and succinate.- Add 5 µL of 0.8 M Malate (2 mM final concentration) and proceed immediately to the next injection. Wash the injection syringe 3 times in water.
- Immediately add 5 µL of 1 M Pyruvate (2.5 mM final concentration) and wait for the respiration to stabilize. Wash the injection syringe 3 times in water.
- Add 10 µL of 0.5 M ADP (2.5 mM final concentration) and wait for the ADP response to stabilize. Wash the injection syringe 3 times in water.
NOTE: Additional ADP (2.5-10 mM) may be required to ensure that the adenylate concentrations are not limiting to respiratory fluxes. - Add 5 µL of 2 M Glutamate (5 mM final concentration) and wait for the respiration to stabilize. Wash the injection syringe 3 times in water.
- Add 5 µL of 4 mM Cytochrome c (10 µM final concentration) and wait for the respiration to stabilize. Wash the injection syringe 3 times in water.
- Add 20 µL of 1 M Succinate (10 mM final concentration) and wait for the respiration to stabilize. Wash the injection syringe 3 times in water.
- Titrate 0.5-1 µL increments of 1 mM FCCP (2-20 µM final concentration) and wait for the respiration to stabilize after each injection, continue until there is no additional increase in respiration. Wash the injection syringe 3 times in 70% ethanol.
- Add 2 µL of 150 µM Rotenone (150 nM-2 µM final concentration) and wait for the respiration to stabilize. Add another 1 µL of Rotenone to insure there is no further inhibition. If there is a decrease in respiration, continue with additional injections until there is no decrease in respiration. Wash the injection syringe 3 times in 70% ethanol.
- Add 2 µL of 125 µM Antimycin A (125 nM-5 µM final concentration) and wait for the respiration to stabilize. Add another 1 µL of Antimycin A to insure there is no further inhibition. If there is a decrease in respiration, continue with additional injections until there is no decrease in respiration. Wash the injection syringe 3 times in 70% ethanol.
- Check the oxygen concentration of the chamber; if the concentration is below 125 µM, reoxygenate to room air or mildly hyperoxygenate to ensure that oxygen does not limit the respiratory flux. Add 5 µL of 0.8 M Ascorbate (2 mM final concentration). Wash the injection syringe 3 times in 70% ethanol.
- Immediately add 10 µL of 0.2 M TMPD (1 mM final concentration) wait for an increase in respiration slow. Wash the injection syringe 3 times in 70% ethanol.
- Add 25 µL of 4 M Sodium Azide (50 mM final concentration) immediately when the respiratory flux of Ascorbate/TMPD plateaus. Wash the injection syringe 3 times in 70% ethanol.
- End the study- click on File, Save and Disconnect. Continue to wash the chamber and syringe following steps 9.2-9.3.
6. ADP sensitivity protocol
- Once the instrument is calibrated and the sample is prepared, remove the stoppers with a twisting motion and aspirate the MiR05 from the chambers (avoiding the membrane exposed on the inside of the chamber). Mix the homogenate well and add 2.25 mL of the homogenate to the chamber. Suppose adding one homogenate to multiple chambers, pipette 1 mL at a time into each chamber while mixing the homogenate to ensure equal tissue distribution. Click on F4 to name and time stamp the event and click on OK.
- Insert the stoppers, and with a twisting motion, slowly close the chamber and allow the respiration to equilibrate (~15-20 min). Fill the stopper central capillary with MiR05 if necessary.
- Analytical determination of succinate-linked mitochondrial ADP sensitivity: Click on F4 with each injection to name and time stamp the events for each chamber in real-time. Throughout the study, select F6 to adjust the O2 concentration and O2 slope neg. scales as needed.
- Add 2 µL of 150 µM Rotenone (150 nM final concentration).
- Immediately add 20 µL of 1 M Succinate (10 mM final concentration) and wait for the respiration to stabilize.
- Titrate ADP by stepwise addition of sub-saturating concentrations until reaching the maximal response rate (VMAX; 2.5-10 mM final concentration).
NOTE: The rate may plateau after injection due to a small change in ADP concentration, thus increase the injection concentration after each plateau and continue with the titration until there is no further increase in respiration even with a fold increase in ADP injection concentration.
7. Recommended optimization experiments
- Determine optimal homogenate and oxygen concentration for protocol.
- Perform SUIT protocol (steps 5.1-5.3) at multiple tissue concentrations (e.g., 30 mg/mL, 20 mg/mL, 10 mg/mL, 5 mg/mL, 2.5 mg/mL, 1 mg/mL, and/or 0.5 mg/mL).
- Select a concentration that maximizes the respiratory flux while limiting the frequency of reoxygenations (no more than 1-2). If more frequent reoxygenations are required, decrease the concentration of the homogenate.
- Determine the optimal number of strokes with the homogenizer.
- Perform SUIT protocol (steps 5.1-5.3) at multiple levels of homogenization (e.g., 5 strokes, 10 strokes, 15 strokes, 20 strokes).
- Since there are insufficient data in the literature to determine a threshold of the percentage increase of cytochrome c with tumor homogenate preparation, choose the preparation with limited cytochrome c response but adequate respiration energized by the substrate(s) of interest.
- Determine the optimal substrate, ADP, uncoupler, and inhibitor concentrations required for quantitative and reproducible respiratory fluxes.
- Perform SUIT protocol (steps 5.1-5.3.9) at the chosen tissue concentration (step 7.1). Titrate each substrate, uncoupler, inhibitor, and ADP until no further response is observed. Titrate the inhibition with sodium azide in separate experiments.
- Add 5 µL of 0.8 M Malate (2 mM final concentration) and proceed immediately to the next injection.
- Titrate 1 µL increments of 1 M Pyruvate and wait for the respiration to stabilize after each injection, continue until there is no additional increase in respiration.
- Titrate 2 µL increments of 0.5 M ADP and wait for the respiration to stabilize after each injection, continue until there is no additional increase in respiration.
- Titrate 1 µL increments of 2 M Glutamate and wait for the respiration to stabilize after each injection, continue until there is no additional increase in respiration.
- Add 5 µL of 4 mM Cytochrome c (10 µM final concentration) and wait for the respiration to stabilize.
- Titrate 5 µL increments of 1 M Succinate and wait for the respiration to stabilize after each injection, continue until there is no additional increase in respiration.
- Titrate 5 µL increments of 0.5 M ADP and wait for the respiration to stabilize after each injection, continue until there is no additional increase in respiration.
- Titrate 0.5 µL increments of 1 mM FCCP and wait for the respiration to stabilize after each injection, continue until there is no additional increase in respiration.
- Titrate 1 µL increments of 150 µM Rotenone and wait for the respiration to stabilize after each injection, continue until there is no decrease increase in respiration.
- Titrate 1 µL increments of 125 µM Antimycin A and wait for the respiration to stabilize after each injection, continue until there is no additional decrease in respiration.
- Add 5 µL of 0.8 M Ascorbate (2 mM final concentration).
- Immediately add 5 µL of 0.2 M TMPD (.5 mM final concentration) wait for increase in respiration slow. In a separate experiment, add 10 µL of 0.2 M TMPD (1 mM final concentration).
- In separate experiments add, 10 µL, 25 µL, 50 µL, and 100 µL of 4 M Sodium Azide (20 mM, 50 mM, 100 mM, 200 mM final concentration, respectively) immediately when the respiratory flux of Ascorbate/TMPD plateaus.
- Choose the substrate and inhibitor concentrations that are saturating within the first injection to improve the timing of the experiment. Use ADP at saturating or sub-saturating concentrations to combine ADP sensitivity evaluations with traditional SUIT protocols.
- Use sub-maximal uncoupler concentrations to demonstrate the dose-responsiveness without inhibition of respiration flux.
- Perform SUIT protocol (steps 5.1-5.3.9) at the chosen tissue concentration (step 7.1). Titrate each substrate, uncoupler, inhibitor, and ADP until no further response is observed. Titrate the inhibition with sodium azide in separate experiments.
8. Data analysis
- SUIT analysis
- Select the steady-state or peak rates from each titration and export for analytical reduction.
- Express the data as pmol O2 per second per mg of tissue (pmol/s/mg).
- Achieve the analytical reduction of PM-L, PM-P, PMG-P, PMGS-P, and PMGS-E by subtracting antimycin A insensitive rate from the respective rate (i.e., steady-state rate obtained after addition of ADP).
NOTE: PM: Pyruvate + Malate; PMG: Pyruvate + Malate + Glutamate; PMGS: Pyruvate + Malate + Glutamate + Succinate; -L:Leak state; -P: Oxidative phosphorylation state, -E: Electron transfer state. - Achieve the analytical reduction of CIV-E by subtracting the sodium azide insensitive rate from the peak ascorbate/TMPD rate.
- Achieve the analytical reduction of PMG-E by subtracting the rotenone insensitive rate from the PMGS-E rate.
- Achieve the analytical reduction of S-E by subtracting the antimycin A insensitive rate from the rotenone insensitive rate.
- Achieve the analytical reduction of cytochrome c control efficiency, a marker of outer membrane intactness, by the following equation:
jc = ((JCHNOc - JCHNO)/JCHNO) x 100
In the equation, jc is the % increase upon addition of cytochrome c, JCHNOc is the oxygen flux after the addition of cytochrome c, and JCHNO is the oxygen flux prior to the addition of cytochrome c.
- ADP sensitivity analysis
- Analytically determine the kinetics for the ADP sensitivity relative to the leak rate of succinate + rotenone (not the rate of the tissue homogenate).
- Plot the O2 flux (Y-axis) against the relative ADP concentration (X-axis). Determine the maximum respiratory velocity (Vmax) as the peak rate achieved over the ADP titration.
- Determine the Michaelis-Menten kinetics using a curve fitting software (PRISM, version 10.1) to reveal the ADP concentration at which 1/2 VMAX is achieved (Apparent KM).
9. Instrumental quality control
- Perform instrumental O2 background over the desired oxygen concentration range (0-600 µM) and zero calibration.
- Complete steps 3.1 and 3.2
- Remove the stoppers and aspirate the 70% ethanol from the chambers (avoiding the membrane exposed on the inside of the chamber). Rinse 4 times with double distilled H2O, and fill the chamber with 2.25 mL of MiR05
- Hyperoxygenate the chambers to ~600 µM oxygen.
- Insert the stoppers slowly to their fully closed position. Siphon off the excess medium ejected through the injection capillary and collected in the well of the stopper and allow the oxygen signal to stabilize (30-45 min).
- To perform oxygen calibration, select the region where both the oxygen concentration and slope are stable, open the calibration window for the respective chamber, and select the R1 mark.
- Prepare the dithionite solution by dissolving 20 mg of sodium hydrosulfite in 0.5 mL of water. Limit the exposure to air as dithionite is oxidized over time with exposure to oxygen.
- Once the oxygen signal is stabilized, inject 1 µL and observe the decline in oxygen concentration. Adjust the potency of the dithionite solution as needed.
- Inject sufficient dithionite solution to lower the oxygen concentration to 450 µM, 300 µM, 225 µM, 150 µM, 75 µM, and 0 µM, respectively. At each injection, allow the oxygen to stabilize and select a mark for the steady-state slope. Once the oxygen concentration is ~0 µM, mark the oxygen concentration.
- To perform instrumental O2 background correction, select the flux/slope window for the respective chamber, select O2 Background Calibration, and click on Calibrate for the marks of interest.
- To perform zero calibration, open the calibration window for the respective chamber, and select the R0 mark attained after dithionite titration.
- Instrument cleaning
- Quickly rinse each chamber with pure water three times. For the first wash, aspirate the homogenate, fill the chamber fully with water, and then aspirate the water. For the second wash, fill the chamber ¾ full of water, insert the stoppers to force water through the injection capillary, aspirate some of the water through the injection capillary and then remove the stoppers to aspirate wash fully.
- Wash the chambers with 70% ethanol for 5 min.
- Over a sink or beaker, clean the stoppers with pure water, 70% ethanol, and 100% ethanol, forcing fluid through the injection capillaries using a wash bottle.
- Quickly rinse each chamber with pure water two times.
- Incubate the chambers with 2 mL of PBS containing ~2 mg of frozen mitochondria, cell lysate, or living fibroblasts for 15 min.
- Wash the chambers with pure water for 5 min two times.
- Wash the chambers with 70% ethanol for 5 min two times.
- Wash the chambers with 100% ethanol for 10 min.
- Fill the chambers with 70% ethanol.
- If running a consecutive experiment, leave the chambers in 70% ethanol for 5 min and then continue to step 3.3.
- If experiments are complete, put lids on the stoppers and turn off the instrument.
- After each use, properly clean the injection syringes and keep the syringes dedicated for specific compound use to prevent carry-over.
- Insert the syringe into the wash fluid and fully submerge the injection needle.
- Draw up the wash fluid into the syringe to the maximum volume.
- Remove the syringe from the wash container, eject the wash fluid into a beaker and then blot the syringe on a paper towel.
- Repeat steps 9.3.1-9.3.3 three times as soon as possible after each use.
Results
Initial studies revealed that EO771 tumors were lowly oxidative and thus required high homogenate concentrations for adequate O2 flux assessment. Optimization experiments were conducted to determine the optimal tissue homogenate concentration range for the study. Tumor homogenates were initially prepared at 40 mg/mL and then linearly diluted. The O2 flux normalized to tissue mass was consistent across concentrations (Figures 1A-D). It was observed that 40 mg/mL resulted in rapid oxygen depletion and was not suitable for experimentation (Figure 1A). The oxygen consumption slowed substantially with 30 mg/mL and 20 mg/mL but still decreased rapidly in a short time in the absence of substrates or ADP (Figure 1B,C). The 10 mg/mL concentration resulted in the optimum oxygen consumption rate (Figure 1D) that would support a longer, 90-min SUIT protocol.
A SUIT protocol was used to evaluate NADH- and succinate-linked OXPHOS and ET, as well as CIV activity (Figure 2A). Pyruvate and malate were added to the tissue homogenate in the absence of ADP to drive leak (L) through NADH. Saturating ADP was then added to drive maximal NADH-linked OXPHOS (P), followed by the addition of glutamate. Cytochrome c was then added to ensure outer membrane integrity; the increase in respiration rate was less than 20% in all samples (Figure 2B). Given the very low response to NADH-linked substrates, cytochrome c release was also assessed in the presence of succinate and rotenone and observed minimal cytochrome c stimulation (Figure 2B). Interestingly, NADH-linked OXPHOS was negligible in EO771 tumors (Figure 2C). Succinate was then added in the presence of pyruvate, malate, and glutamate to stimulate electron flow through succinate dehydrogenase. FCCP was then titrated to drive maximal electron flow (E), which revealed that in EO771 tumors, phosphorylation rather than oxidation was limiting to respiration (Figure 2C). Rotenone and antimycin A were subsequently titrated to inhibit complex I and complex III, respectively. Ascorbate and TMPD were then added to drive maximal electron flow through CIV, which is then inhibited by sodium azide. Table 1 illustrates analytical reduction equations of the raw data (Table 2) to quantitate the respiratory parameters plotted in Figure 2C. Overall, the tumor homogenate respiratory profiles (Figure 2C) are similar to those of non-implanted digitonin-permeabilized EO771 cells (Figure 2D) with the exception of diminished maximal electron transfer supported by N- and S-linked substrates in the tumor.
Since NADH-linked respiration was negligible, the respiratory kinetics of succinate were further evaluated by stepwise titrations of sub-saturating ADP until the maximal rate (VMAX) was achieved (Figure 3A,3B). The half-maximal concentration (KM) of ADP in the presence of succinate + rotenone was 37.5 µM, whereas the VMAX was ~10.5 pmol/s/mg (Figure 3C). Thus, despite relatively poor oxidation rates, EO771 tumors were highly sensitive to ADP and sustained ATP synthesis at relatively low ADP concentrations.
Selecting appropriate regions of the raw data for extraction is critical for the reproducibility of experiments and accurate quantification. For cytochrome c, a mark needs to be selected at the steady-state immediately prior to injection (Figure 4A, mark 1). There is often an initial injection artifact that can be followed by a period of time (about 5-10 min) where the O2 flux is not steady. Evaluation of cytochrome c efficiency is made by making an additional selection once the O2 flux has stabilized (Figure 4A, mark 2). Selections after the addition of substrates, ADP, or most inhibitors are also made after the injection artifact and once the O2 flux has stabilized (Figure 4B). The selection used to determine maximal uncoupled respiration is made at the peak increase achieved during titration of FCCP, which is often not the last injection made (Figure 4C). The selection for TMPD is made after both ascorbate and TMPD have been added and at the peak increase in respiration (Figure 4D, mark 1). Just after this peak, the inhibitor, sodium azide, is added, which rapidly decreases respiration but also often has an injection artifact lower than the inhibited respiration rate (Figure 4D). The inhibitor mark is made immediately after the injection artifact (Figure 4D, mark 2). The O2 flux will typically not stabilize and continue to decrease.
Table 1: Respiratory notation and analytical derivation. Please click here to download this Table.
Table 2: Sample and respiratory characteristics of luminal B mammary tumor homogenates. Please click here to download this Table.

Figure 1: Optimization of tumor homogenate concentration. O2 Flux (red) and O2 Concentration (blue) in mammary tumor homogenates prepared at (A) 40 mg/mL, (B) 30 mg/mL, (C) 20 mg/mL, and (D) 10 mg/mL. Thom: Tissue homogenate respiration. Please click here to view a larger version of this figure.
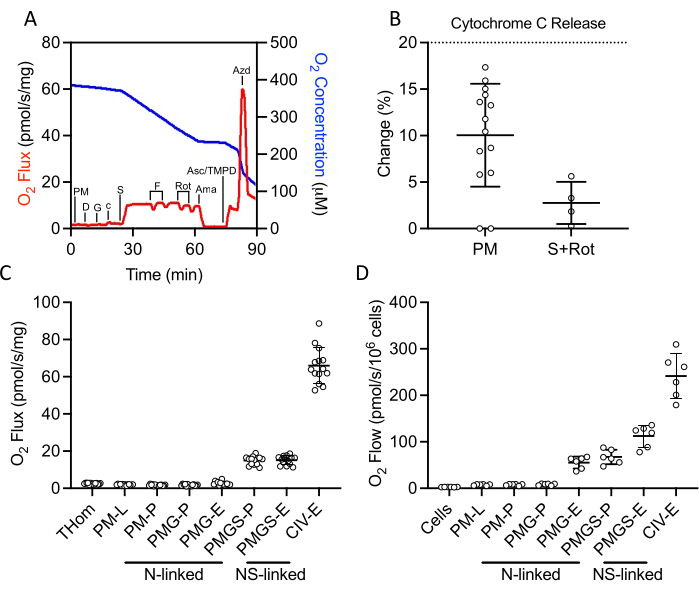
Figure 2: Evaluation of OXPHOS and ET capacity by high-resolution respirometry in freshly excised tumor homogenates. (A) Representative plot of oxygen consumption (red) and concentrations (blue) over the course of a substrate, inhibitor, uncoupler protocol. PM: Pyruvate + Malate, D: ADP, G: Glutamate, c: Cytochrome c, S: Succinate, F: FCCP, Rot: Rotenone, Ama: Antimycin A, Asc/TMPD: Ascorbate/Tetramethyl-p-phenylenediamine. (B) Percent increase in O2 flux upon addition of cytochrome c. (C-D) Respiration supported by malate, pyruvate, glutamate, and succinate in the presence of ADP, FCCP, and ascorbate/TMPD in (C) EO771-derived tumor homogenates and (D) non-implanted EO771 digitonin-permeabilized cells. Thom: Tissue homogenate respiration; PM: Pyruvate + Malate; PMG: Pyruvate + Malate + Glutamate; PMGS: Pyruvate + Malate + Glutamate + Succinate; CIV: Complex IV; -L: Leak state; -P: Oxidative phosphorylation state, -E: Electron transfer state; N-linked: O2 flux supported by defined NADH-generating substrate combinations; NS-linked: O2 flux supported by the convergence of defined NADH-generate substrate combinations and succinate. Please click here to view a larger version of this figure.
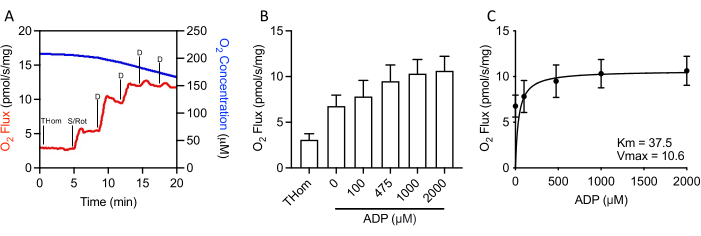
Figure 3: EO771 mammary tumors displayed high ADP (adenosine 5′-diphosphate) sensitivity. (A) Representative plot of oxygen consumption (red) and concentrations (blue) throughout an S-linked ADP titration protocol. Thom: Tissue homogenate respiration; S/Rot: Succinate/Rotenone; D: ADP. (B) Respiration supported by succinate in the presence of rotenone and increasing concentrations of ADP (0 µM ADP = S/Rot-L). (C) Maximal rate (VMAX) and half-maximal concentration (KM) of ADP in the presence of succinate + rotenone. Please click here to view a larger version of this figure.
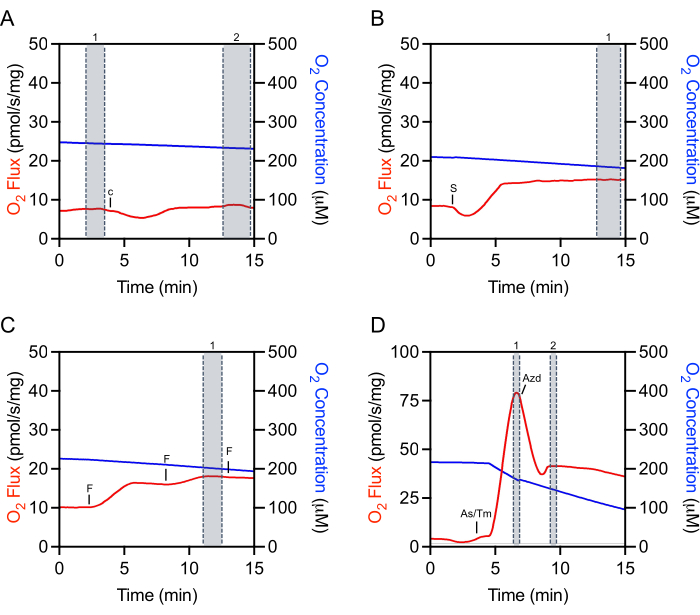
Figure 4: Representative tracing illustrating mark selection of raw O2 fluxes for data extraction. (A) Cytochrome c selection: selection number 1 before the Cytochrome c injection and selection number 2 after the injection when the O2 flux has stabilized. c Cytochrome c. (B) Substrate, ADP, and inhibitor selection: selection number 1 after the injection (succinate in this representative plot) where the O2 flux has stabilized. S: Succinate. (C) Uncoupler selection: selection number 1 at the peak increase in respiration during the uncoupler titration. In this representative FCCP titration plot, the third injection slightly decreases respiration and thus is not used for selection. F: ACCP. (D) TMPD selection: selection number 1 at the peak increase of respiration after the ascorbate and TMPD injections. Sodium azide selection: selection number 2 after the acute injection artifact when the respiration initially decreases. As/Tm: Ascorbate/TMPD; Azd: Azide. Please click here to view a larger version of this figure.
Discussion
Approaches to evaluating mitochondrial respiration in cancer have largely been limited to in vitro models13,14,15,16. Some success has been achieved in measuring mitochondrial respiration in tumors using chemical permeabilization6,7,17, but there is no uniform, gold-standard approach that can be universally applied and compared across tumor types. Additionally, a lack of consistent data analysis and reporting has limited data generalizability and reproducibility. The method outlined herein provides a simple, relatively quick approach to measure mitochondrial respiration18 in mitochondrial preparations from freshly excised solid tumor specimens. Tumors were grown from orthotopically implanted murine Luminal B, ERα-negative EO771 mammary cancer cells19.
Diligence and care with tissue handling will greatly improve the accuracy and normalization of the oxygen consumption rates. The tissue and mitochondria can be easily damaged if the sample is not kept cold, is not consistently submerged in preservation media, or is overly handled, resulting in suboptimal routine and OXPHOS rates. Additionally, accurate wet weight of the homogenized tissue is of critical importance as this is the primary normalization method. Other normalization methods may be considered, such as total protein or mitochondrial specific markers, such as citrate synthase activity20. Additionally, tissue heterogeneity will need to be addressed, with decisions about tumor regions to include in experiments made a priori. Necrotic, fibrotic, and connective tissue may not homogenize and/or respire well and should be avoided unless intentionally assaying these tumor regions. Notably, the tumor may be very sticky depending on the type and excision region, making accurate weighing and transfer more challenging. The number of strokes used for homogenizations should be optimized to ensure complete preparation of the mitochondria while mitigating damage to the outer mitochondrial membranes.
For improved accuracy and reproducibility, we recommend optimization experiments be performed for the number of strokes for homogenate preparation, tissue concentration, and substrate, uncoupler, inhibitor concentrations. Studies can compare the different number of strokes and how they correspond with response to the addition of cytochrome c within the study as well as the maximal mitochondrial respiratory capacity 21. Although there is a general acceptance that less cytochrome c response is better, as an increase in oxygen consumption after the addition of cytochrome c can indicate damage to the outer mitochondrial membrane, there is no gold-standard as to what this threshold is for every tissue and should be experimentally investigated to ensure the tissue is not being overworked or underprepared. In this tumor tissue, it was found that a cytochrome c response under ~30% did not impair respiratory function. Cytochrome c use becomes critical for accurate quantification of respiratory capacity if the test is positive. In this case, the addition replenishes endogenous cytochrome c, which, if depleted, will cause an underestimation of the respiratory rates.
Tissue concentration titration experiments can be performed over a range of feasible concentrations and, ideally, would be done with SUITs that will be investigated during the study. Respiratory capacity will vary by tumor type and composition. Thus, tumors dense with mitochondria or high respiratory capacity will require lower concentrations (0.5-5 mg/mL). Tumors with few mitochondria or low respiratory capacity will require higher concentrations (7-12 mg/mL). Additionally, SUITs that are long or have highly consumed substrates may need less tissue to prevent reoxygenation of the chamber or ADP limitation. Some tissues will have a linear relationship in oxygen consumption, whereas others will show improved sensitivity and maximal oxidation at certain concentration ranges. The chosen tissue concentration should be optimized to maximize oxygen flux while limiting the number of reoxygenation events. Additionally, it is often better to overestimate the need or aim for the higher end of the concentration range. The inhibitors, which are essential to the quantitation of respiratory fluxes, are more precise when used in larger pools of mitochondria.
Another essential consideration is the concentration of the drugs that are used during the protocols. Changes in homogenate concentration may alter the concentrations of substrates, uncouplers, and inhibitors required for maximal response. Thus, once the optimal concentration range is chosen, an experiment testing the doses required for the SUIT protocol should be performed. Additional ADP can be added to ensure that adenylate concentrations are not limiting to respiratory fluxes. Chemical uncouplers such as FCCP or CCCP will inhibit respiration at higher concentrations22. As such, it is essential to titrate in small amounts to reveal the maximal achieved rate. Inhibitors, such as rotenone and antimycin A, are best used when saturated within the first injection. While optimal concentrations were determined in preliminary experiments, we have also observed treatment-related differences in response to inhibitors and thus often add one additional injection of inhibitors to demonstrate maximal inhibition as the resultant rates serve as the basis for quantification. Chemical inhibition of Ascorbate/TPMD is essential for accurate analytical reduction as TMPD undergoes autooxidation23. We controlled for auto-oxidation of ascorbate/TMPD/cytochrome c through the addition of sodium azide, an established CIV inhibitor. For the Km studies, the addition of rotenone in the presence of succinate alone prevents oxaloacetate accumulation which can inhibit succinate dehydrogenase activity at low concentrations24. The volume and concentration of ADP are highly dependent on the sensitivity of the mitochondria to the prevailing substrate combination. Mitochondrial preparations that are highly sensitive to ADP will require lower starting concentrations. Additionally, validated chemicals and proper drug preparation with attention to pH, sensitivity to light if applicable, and storage temperature are essential for successful experiments.
Instrument setup and routine care are of critical importance for the success of these experiments. Adequate and proper cleaning of the chambers is essential for reproducibility and prevention of biological, protein, inhibitor, or uncoupler contamination. Clark-type electrodes and O2k systems utilize glass reaction chambers which is a significant cost advantage to plate-based systems which rely on consumables. However, the glass chambers must be vigorously cleaned and can be a source of inhibitor contamination in subsequent studies. Incubation with mitochondria-rich specimens during the washing process (isolated heart or liver mitochondria, for example) can reduce the risk of experimental contamination and is recommended in addition to dilution and alcohol-based washing procedures. If consecutive studies are run, cleaning with ethanol and mitochondria minimizes the possibility of inhibitor contamination. Calibration of the oxygen sensor is recommended prior to each experiment to obtain accurate measurements of respiration relative to the prevailing partial pressure of oxygen. If multiple calibrations are not feasible, one calibration a day may be sufficient if the oxygen concentration remains stable and consistent after the washing procedure.
The procedures outlined above leverage the Oroboros O2k instrument for measuring oxygen consumption in tumor tissue within 4 hours of tumor excision using previously designed and optimized preservation solution and respiration media25,26,27. Multiple parameters in this protocol can be modified for subsequent applications. The instrument setup and calibration, the homogenizers used for tissue preparation, and optimal homogenate and chamber oxygen concentration can all be adapted for use on other instruments with oxygen monitoring potential. For example, the chambers were slightly overfilled when adding homogenate, and thus when the chamber is fully closed, the chamber capillary remains full. This will consume some oxygen in the chamber, but with optimization of sample concentration, we can account for this consumption in determining what oxygen level to start with. Alternatively, the sample can be allowed to equilibrate at ambient oxygen before the chamber is closed, but this will often increase the amount of time before the experiment starts and delay the addition of substrates. While the homogenizers used in this protocol are widely accessible, other commercial homogenization techniques could be employed, such as a tissue shredder or automated homogenizer28.
Additionally, the tissue preparation and instrument procedures can be utilized with a number of different SUITs to study respiratory control by a diversity of coupling and pathway control states29. These SUIT protocols have been developed to measure functional capacity, and thus, the contribution of potential endogenous substrates has no impact on the capacity measurement. We analytically account for non-mitochondrial oxygen consumption and/or residual consumption of the homogenate through subtraction of the antimycin A-rotenone, or sodium azide insensitive rates, as appropriate. Mitochondria can remain viable in BIOPS or similarly constructed preservation solutions for extended periods of time (>24 h) depending on tissue type and intactness30,31. Studies can be carried out in advance to determine the temporal storage limits as OXPHOS of certain substrates may have different limitations. This is essential if the experiment cannot be performed within several hours of tissue excision/biopsy. 37°C is an optimal and physiological temperature for the evaluation of respiratory function in most mammalian systems. However, if the assay temperature appears to interfere with evaluation32, comparative studies may be conducted across a wide temperature range (25-40 °C) to ensure adequate responsiveness. Instrumental constraints may limit the ability to conduct such studies.
Major limitations of the above-described method are 1) the potential for damage to mitochondria through mechanical homogenization, 2) presence of ATPases or other sub-cellular biochemicals in homogenate preparations that can interfere with simultaneous determination of ATP or other variables of interest and may require additional correction methods or inhibitor use33, and 3) evaluation of many samples and/or multiple SUITs per sample is time-consuming as one instrument can accommodate two experiments at a time and requires cleaning and set up in-between successive experiments. Optimization experiments and consistent preparation of samples can minimize substantial mitochondrial damage that would contribute to inconsistent data.
The significance of the method with respect to existing/alternative methods is improved feasibility compared with the amount of starting material, challenge of isolating mitochondria, or technical challenge in permeabilizing tissue. Preparation of homogenates is faster, oxygen is not nearly as limiting, and is less susceptible to variability between personnel compared with permeabilized tissue. Importantly, nearly all sample types are suitable for homogenate preparation which allows for comparative analysis across tissues. High-resolution respirometry is the gold-standard measurement of mitochondrial OXPHOS and ET. The application of this method in pre-clinical and clinical cancer research has the capacity to expand current in vitro investigations to ex vivo studies. Furthermore, it offers potential applications in clinical and diagnostic settings.
Disclosures
The authors have no conflicts of interest related to this work.
Acknowledgements
We thank the Pennington Biomedical Research Center Comparative Biology Core staff for animal care. This research was supported in part by the National Institute of Health grants U54GM104940 (JPK) and KL2TR003097 (LAG). All experiments and procedures involving animals were approved by the Pennington Biomedical Research Center Institutional Animal Care and Use Committee.
Materials
| Name | Company | Catalog Number | Comments |
| 2-(N-Morpholino)ethanesulfonic acid hydrate | Sigma-Aldrich | M8250 | |
| Adenosine 5′-diphosphate sodium salt | Sigma-Aldrich | A2754 | |
| Adenosine 5'-triphosphate disodium salt hydrate | Sigma-Aldrich | A2383 | |
| Amphotericin B | Gibco | 15290018 | |
| Antimycin A | Sigma-Aldrich | A8674 | |
| Ascorbate | Sigma-Aldrich | A4544 | |
| Bovine serum albumin, fraction V, heat shock, fatty acid free | Sigma-Aldrich | 3117057001 | Roche |
| BD 50 mL Luer-Lok Syringe | Fisher Scientific | 13-689-8 | |
| BD Vacutainer General Use Syringe Needles | Fisher Scientific | 23-021-020 | |
| Calcium carbonate | Sigma-Aldrich | C4830 | |
| Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone | Sigma-Aldrich | C2920 | |
| Cytochrome c from equine heart | Sigma-Aldrich | C2506 | |
| Datlab 7.4 software | Oroboros Instruments | ||
| Dimethylsulfoxide | Amresco | N182 | |
| Dithiothreitol | Sigma-Aldrich | D0632 | |
| D-Sucrose | Sigma-Aldrich | S7903 | |
| Dumont # 5 Forceps | Fine Science Tools | 11251-30 | Dumoxel, autoclavable |
| Dumont # 7 Forceps | Fine Science Tools | 11271-30 | Dumoxel, autoclavable |
| Digital Calipers 150 mm/6 in | World Precision Instruments | 501601 | |
| EO771 cells | CH3 BioSystems | SKU: 94APV1-vial-prem | Pathogen Tested |
| Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid | Sigma-Aldrich | E4378 | |
| Female C57BL/6J mice | Jackson Laboratory | Stock #000664 | |
| HEPES | Sigma-Aldrich | H4034 | |
| Imidazole | Sigma-Aldrich | 56750 | |
| Kimwipes | Fisher Scientific | 34120 | |
| L-(−)-Malic acid | Sigma-Aldrich | G1626 | |
| Lactobionic acid | Sigma-Aldrich | L2398 | |
| Malate | Sigma-Aldrich | M6413 | |
| Matrigel Matrix | Corning | 354248 | |
| MgCl·6H2O | Sigma-Aldrich | M2670 | |
| Microsyringes | Hamilton | 87919, 80383, 80521, 80665, 80765, 80865, 87943 | |
| N,N,N′,N′-Tetramethyl-p-phenylenediamine | Sigma-Aldrich | T7394 | |
| Oxygraph-2k | Oroboros Instruments | 10023-03 | |
| Oxygraph-2k FluoRespirometer | Oroboros Instruments | 10003-01 | |
| PBS | Gibco | 10010023 | |
| Penicillin-Streptomycin | Gibco | 15140122 | |
| Phosphocreatine disodium salt hydrate | Sigma-Aldrich | P7936 | |
| Potassium hydroxide | Sigma-Aldrich | P1767 | |
| Potassium phosphate monobasic | Sigma-Aldrich | P5655 | |
| Rotenone | Sigma-Aldrich | R8875 | |
| RPMI 1640 | Gibco | 21875034 | |
| Sodium azide | Sigma-Aldrich | S2002 | |
| Sodium pyruvate | Sigma-Aldrich | P5280 | |
| Succinate (disodium) | Sigma-Aldrich | W327700 | |
| Taurine | Sigma-Aldrich | T0625 | |
| Whatman Filter Paper, grade 5 | Sigma-Aldrich | 1005-090 | |
| Wheaton Tenbroeck Tissue Grinder, 7 mL | Duran Wheaton Kimble | 357424 | |
| Straight Tip Micro Dissecting Scissors | Roboz | RS-5914SC | |
| Non-Safety Scalpel No. 11 | McKesson | 1029065 | |
| BD Precision Glide Needle 27 G x 1/2 | Becton, Dickinson and Company | 305109 | |
| BD Precision Glide Needle 18 G x 1 | Becton, Dickinson and Company | 305195 | |
| BD 1mL Slip Tip Syringe | Becton, Dickinson and Company | 309659 | |
| Pyrex Reusable Petri Dish, 60 mm | Thermo Fisher Scientific | 316060 | |
| Rodent Very High Fat Diet, 60% kcal from fat, 20% kcal from protein, and 20% kcal from carbohydrate | Research Diet | D12492 | |
| Pyrex Watch Glass, 100 mm | Thermo Fisher Scientific | S34819 |
References
- DeBerardinis, R. J., Chandel, N. S. Fundamentals of cancer metabolism. Science Advances. 2 (5), 1600200(2016).
- Bajzikova, M., et al. Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metabolism. 29 (2), 399-416 (2019).
- Martínez-Reyes, I., et al. Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature. 585 (7824), 288-292 (2020).
- Picard, M., et al. Mitochondrial structure and function are disrupted by standard isolation methods. PloS One. 6 (3), 18317(2011).
- Saks, V. A., et al. Bioenergetics of the Cell: Quantitative Aspects. Saks, V. A., et al. , Springer US. 81-100 (1998).
- Kaambre, T., et al. Metabolic control analysis of cellular respiration in situ in intraoperational samples of human breast cancer. Journal of Bioenergetics and Biomembranes. 44 (5), 539-558 (2012).
- Koit, A., et al. Mitochondrial respiration in human colorectal and breast cancer clinical material is regulated differently. Oxidative Medicine and Cellular Longevity. 2017, 1372640(2017).
- Holland, O. J., et al. Changes in mitochondrial respiration in the human placenta over gestation. Placenta. 57, 102-112 (2017).
- Pesta, D., Gnaiger, E. High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods in Molecular Biology. 810, 25-58 (2012).
- Makrecka-Kuka, M., Krumschnabel, G., Gnaiger, E. High-resolution respirometry for simultaneous measurement of oxygen and hydrogen peroxide fluxes in permeabilized cells, tissue homogenate and isolated mitochondria. Biomolecules. 5 (3), 1319-1338 (2015).
- Kondrashova, M. N., et al. Preservation of native properties of mitochondria in rat liver homogenate. Mitochondrion. 1 (3), 249-267 (2001).
- Pecinová, A., Drahota, Z., Nůsková, H., Pecina, P., Houštěk, J. Evaluation of basic mitochondrial functions using rat tissue homogenates. Mitochondrion. 11 (5), 722-728 (2011).
- Wu, M., et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. American Journal of Physiology-Cell Physiology. 292 (1), 125-136 (2007).
- Hlozková, K., Starková, J. Assessment of the metabolic profile of primary leukemia cells. Journal of Visualized Experiments: JoVE. (141), e58426(2018).
- Zhang, J., Zhang, Q. Cancer Metabolism: Methods and Protocols. Haznadar, M. , Springer. New York. 353-363 (2019).
- Wigner, P., Zielinski, K., Labieniec-Watala, M., Marczak, A., Szwed, M. Doxorubicin-transferrin conjugate alters mitochondrial homeostasis and energy metabolism in human breast cancer cells. Scientific Reports. 11 (1), 4544(2021).
- Schöpf, B., et al. Oxidative phosphorylation and mitochondrial function differ between human prostate tissue and cultured cells. The FEBS Journal. 283 (11), 2181-2196 (2016).
- Ye, F., Hoppel, C. L. Measuring oxidative phosphorylation in human skin fibroblasts. Analytical Biochemistry. 437 (1), 52-58 (2013).
- Le Naour, A., et al. EO771, the first luminal B mammary cancer cell line from C57BL/6 mice. Cancer Cell International. 20, 328(2020).
- Gnaiger, E., et al. Mitochondrial physiology. Bioenergetics Communications. 1, 44(2020).
- Hughey, C. C., Hittel, D. S., Johnsen, V. L., Shearer, J. Respirometric oxidative phosphorylation assessment in saponin-permeabilized cardiac fibers. Journal of Visualized Experiments: JoVE. (48), e2431(2011).
- Dranka, B. P., et al. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radical Biology and Medicine. 51 (9), 1621-1635 (2011).
- Munday, R. Generation of superoxide radical, hydrogen peroxide and hydroxyl radical during the autoxidation of N,N,N',N'-tetramethyl-p-phenylenediamine. Chemico-Biological Interactions. 65 (2), 133-143 (1988).
- Moser, M. D., Matsuzaki, S., Humphries, K. M. Inhibition of succinate-linked respiration and complex II activity by hydrogen peroxide. Archives of Biochemistry and Biophysics. 488 (1), 69-75 (2009).
- Veksler, V. I., Kuznetsov, A. V., Sharov, V. G., Kapelko, V. I., Saks, V. A. Mitochondrial respiratory parameters in cardiac tissue: a novel method of assessment by using saponin-skinned fibers. Biochimica et Biophysica Acta. 892 (2), 191-196 (1987).
- Gnaiger, E., et al. Mitochondria in the Cold. Life in the Cold. Heldmaier, G., Klingenspor, M. , Springer. Berlin, Heidelberg. 431-442 (2000).
- Doerrier, C., et al. High-resolution fluorespirometry and OXPHOS protocols for human cells, permeabilized fibers from small biopsies of muscle, and isolated mitochondria. Methods in Molecular Biology. 1782, 31-70 (2018).
- Rohlenova, K., et al. Selective disruption of respiratory supercomplexes as a new strategy to suppress Her2(high) breast cancer. Antioxidants & Redox Signaling. 26 (2), 84-103 (2017).
- Gnaiger, E. Mitochondrial pathways and respiratory control. Bioenergetics Communications. 5th ed. , (2020).
- Barksdale, K. A., et al. Mitochondrial viability in mouse and human postmortem brain. FASEB Journal. 24 (9), 3590-3599 (2010).
- Underwood, E., Redell, J. B., Zhao, J., Moore, A. N., Dash, P. K. A method for assessing tissue respiration in anatomically defined brain regions. Scientific Reportys. 10 (1), 13179(2020).
- Jorgensen, L. B., Overgaard, J., Hunter-Manseau, F., Pichaud, N. Dramatic changes in mitochondrial substrate use at critically high temperatures: a comparative study using Drosophila. Journal of Experimental Biology. 224, Pt 6 (2021).
- Salin, K., et al. Simultaneous measurement of mitochondrial respiration and ATP production in tissue homogenates and calculation of effective P/O ratios. Physiological Reports. 4 (20), 13007(2016).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved