A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Layer Microdissection of Tricuspid Valve Leaflets for Biaxial Mechanical Characterization and Microstructural Quantification
In This Article
Summary
This protocol describes the biaxial mechanical characterization, polarized spatial frequency domain imaging-based collagen quantification, and microdissection of tricuspid valve leaflets. The provided method elucidates how the leaflet layers contribute to the holistic leaflet behaviors.
Abstract
The tricuspid valve (TV) regulates the unidirectional flow of unoxygenated blood from the right atrium to the right ventricle. The TV consists of three leaflets, each with unique mechanical behaviors. These variations among the three TV leaflets can be further understood by examining their four anatomical layers, which are the atrialis (A), spongiosa (S), fibrosa (F), and ventricularis (V). While these layers are present in all three TV leaflets, there are differences in their thicknesses and microstructural constituents that further influence their respective mechanical behaviors.
This protocol includes four steps to elucidate the layer-specific differences: (i) characterize the mechanical and collagen fiber architectural behaviors of the intact TV leaflet, (ii) separate the composite layers (A/S and F/V) of the TV leaflet, (iii) carry out the same characterizations for the composite layers, and (iv) perform post-hoc histology assessment. This experimental framework uniquely allows the direct comparison of the intact TV tissue to each of its composite layers. As a result, detailed information regarding the microstructure and biomechanical function of the TV leaflets can be collected with this protocol. Such information can potentially be used to develop TV computational models that seek to provide guidance for the clinical treatment of TV disease.
Introduction
The TV is located between the right atrium and right ventricle of the heart. Throughout the cardiac cycle, the TV regulates the unidirectional blood flow via cyclic opening and closing of the TV anterior leaflet (TVAL), the TV posterior leaflet (TVPL), and the TV septal leaflet (TVSL). These leaflets are complex and have four distinct anatomical layers—the atrialis (A), the spongiosa (S), the fibrosa (F), and the ventricularis (V)—with unique microstructural constituents. The elastin fibers in the atrialis and ventricularis help restore the tissue to its undeformed geometry after mechanical loading1. In contrast, the fibrosa contains a dense network of undulated collagen fibers that contribute to the load-bearing capacity of the leaflets2. Mainly consisting of glycosaminoglycans, the spongiosa has been hypothesized to enable shearing between leaflet layers during heart valve function3. While all three leaflet types have the same anatomical layers, there are variations in the thicknesses of the layers and constituent ratios that have implications for leaflet-specific mechanical behaviors.
Researchers have explored the properties of the TV leaflets using planar mechanical characterizations, histomorphological assessments, and optical characterizations of the collagen fiber architecture. For example, planar biaxial mechanical characterizations seek to emulate physiological loading by applying perpendicular displacements to the tissue and recording the associated forces. The resulting force-displacement (or stress-stretch) observations have revealed that all three TV leaflets exhibit nonlinear, direction-specific mechanical behaviors with more apparent leaflet-specific responses in the radial tissue direction4,5,6. These leaflet-specific behaviors are believed to stem from differences in the microstructural properties observed using standard histological techniques6,7. Further, second harmonic generation imaging6, small-angle light scattering8, and polarized spatial frequency domain imaging7 (pSFDI) aim to understand these microstructural properties and have shown leaflet-specific differences in the collagen fiber orientation and fiber crimp that have implications for the observed tissue-level mechanical behaviors. These studies have significantly advanced our understanding of the tissue microstructure and its role in tissue-level behaviors. However, much remains to be addressed in experimentally connecting the tissue mechanics and the underlying microstructure.
Recently, this laboratory performed mechanical characterizations of the TV leaflet layers separated into two composite layers (A/S and F/V) using a microdissection technique9. That earlier work highlighted differences in the mechanical properties of the layers and helped provide insight into how the layered microstructure contributes to the tissue mechanical behaviors. Although this investigation improved our understanding of the TV leaflet microstructure, the technique had several limitations. First, the properties of the composite layers were not directly compared to the intact tissue, leading to a lack of complete understanding of the mechanics-microstructure relationship. Second, the collagen fiber architecture of the composite layers was not examined. Third, only the layers of the TVAL were investigated due to difficulties with collecting the composite layers from the other two TV leaflets. The method described herein provides a holistic characterization framework that overcomes these limitations and provides complete characterizations of the TV leaflets and their composite layers.
This paper describes the microdissection technique that separates the three TV leaflets into their composite layers (A/S and F/V) for biaxial mechanical and microstructural characterizations10,11,12. This iterative protocol includes (i) biaxial mechanical testing and pSFDI characterization of the intact leaflet, (ii) a novel and reproducible microdissection technique to reliably obtain the composite TV layers, and (iii) biaxial mechanical testing and pSFDI characterization of the composite TV layers. The tissue was exposed to biaxial tensile loading with various force ratios for mechanical testing. Then, pSFDI was used to determine the collagen fiber orientation and alignment at various loaded configurations. pSFDI preserves the native collagen fiber architecture, allows load-dependent analysis, and circumvents the typical need to fix or clear tissue for collagen fiber architecture analysis, such as in second harmonic generation imaging or small-angle light scattering. Finally, the tissues were prepared using standard histology techniques to visualize the tissue microstructure. This iterative and holistic framework allows for the direct comparison of the mechanical and microstructural properties of the TV leaflet to its composite layers.
Access restricted. Please log in or start a trial to view this content.
Protocol
All methods described herein were approved by the Institutional Animal Care and Use Committee at the University of Oklahoma. Animal tissues were acquired from a USDA-approved slaughterhouse.
1. Biaxial mechanical characterization
- Tissue preparation
- Retrieve a TV leaflet from the freezer, razor blades, a surgical pen, forceps, a pipette with deionized (DI) water, and a cutting mat. Thaw the TV leaflet using 2-3 drops of room-temperature DI water.
NOTE: DI water is used instead of phosphate-buffered saline (PBS) to avoid any PBS-induced difficulties for the layer microdissection. - Lay the specimen flat on the cutting mat with the ventricularis layer (i.e., the surface with the chordae insertions) facing upwards. Position the specimen so that the radial direction aligns with the Y-direction and the circumferential direction aligns with the X-direction.
NOTE: The circumferential direction is oriented along the circumference of the valve. - Examine the chordae insertion locations of the tissue. Determine an area, ideally ~12 x 12 mm, with the least amount of large chordae insertions while avoiding extremely thin (i.e., transparent) areas (Figure 1).
- Flip the specimen over so the atrial surface (i.e., the surface with no chordae insertions) is facing upwards. Ensure the circumferential and radial leaflet directions remain aligned with the X- and Y-axes, respectively.
- Cut a square 12 x 12 mm specimen from the leaflet tissue that avoids the large chordae insertions or thin areas identified in step 1.1.3. Remove the trimmed portions of the leaflet tissue with the forceps and place them into a waste container.
- If it is not possible to completely avoid large chordae insertions, cut the tissues so that they are along the edge of the square specimen. Avoiding chordae insertions is important as it helps prevent issues for the later microdissection.
- Use a surgical pen to place a small dot in the top right corner to track the specimen's orientation. Allow the ink to dry for approximately 30 s.
- Flip the specimen with the ventricular surface (i.e., the surface with chordal insertions) facing upwards. Trim the chordal attachments on the back of the tissue by stretching the chordae from the leaflet and using a razor blade to cut near its insertion location. Flip the specimen again so the atrial surface (i.e., the smooth surface) is facing upwards.
- Retrieve a TV leaflet from the freezer, razor blades, a surgical pen, forceps, a pipette with deionized (DI) water, and a cutting mat. Thaw the TV leaflet using 2-3 drops of room-temperature DI water.
- Thickness measurement
- Retrieve a noncontact laser displacement sensor. Zero the displacement sensor on a flat section of the cutting mat near the trimmed tissue.
CAUTION: Do not directly shine the laser into the eyes. - Position the laser over the central region of the specimen. Remove air trapped under the leaflet surface as it can cause measurement errors. To release trapped air, either use tweezers to push the bubble to the edge of the tissue or lift one corner of the tissue.
- Record the thickness shown on the displacement sensor's display. Repeat two more measurements in other locations of the specimen.
- Compute the average leaflet thickness using the three measurements recorded in the previous step. Use this value when creating the biaxial mechanical characterization protocols.
- Retrieve a noncontact laser displacement sensor. Zero the displacement sensor on a flat section of the cutting mat near the trimmed tissue.
- Biaxial tester setup and tissue mounting
- Prepare a DI water bath at 37 °C, following the testing system's guidelines, to ensure the temperature under in vivo physiologic conditions.
- Retrieve forceps, the tissue specimen, mounting hardware, a fine-tipped tool, liquid cyanoacrylate glue, and black-painted glass beads (diameter: 300-500 µm).
NOTE: Mounting hardware includes the tines, the mounting bridge, and the mounting rubber. - Mount the tissue specimen to the testing system. Ensure the circumferential direction of the tissue aligns with the X-direction, which can be assisted by the dot previously placed in step 1.1.6.
NOTE: The tines used here should be evenly spaced across the entire tissue edge length. The effective edge length is set to be 10 mm for the intact tissue and >3.3 mm for the composite layers.
- Fiducial marker placement
- Identify the central one-third square region of the mounted tissue. Use the approximate corners of this area for the fiducial marker placement.
- Place glass beads in an open-faced weighing boat and create a small pool of liquid cyanoacrylate glue in a separate weigh boat. Coat the top of the fine-tipped tool with a small amount of glue. Dab excess glue on the side of the weigh boat.
- Create one corner of the central one-third square array by gently pressing the glue-coated tip onto the tissue. Using forceps, grab a glass bead and carefully place it on top of the glue dot. Use the camera of the biaxial testing device for help with the bead placement.
- Repeat steps 1.4.2 and 1.4.3 for three additional beads until the square array is completed. Ensure the beads are securely attached, and their respective glue dots are not touching or sticking together. Dry the glue before lowering the tissue into the water bath.
- If the beads are stuck together, wait for the glue to dry, then use the forceps to gently grasp the bead or glue and pull it off the tissue.
NOTE: The glue and bead(s) should come off, allowing bead placement to be reattempted.
- If the beads are stuck together, wait for the glue to dry, then use the forceps to gently grasp the bead or glue and pull it off the tissue.
- Preconditioning
- Create a force-controlled preconditioning protocol, in which the tissue with edge length and thickness will undergo six repetitions of equibiaxial loading to a peak membrane tension Tpeak of 40 N/m with a preload of 3% × Tpeak10 and stretch and recovery times of 30 s each.
- Construct an arbitrary testing directory that will temporarily store the data for future calculations. Set the loading rate to be 4.42 N/m.
- Construct a new testing parameter set with the name Preconditioning0. Set the X-axis and Y-axis control modes to force and set the control functions to step. Define the load magnitude to be the force associated with Tpeak, i.e., fpeak = Tpeak · L. Define the preload magnitude as 3% of fpeak for the first repetition only and define both the stretch duration and recovery duration as 30 s. Define the number of repetitions as 10.
NOTE: The computed peak first Piola-Kirchhoff stress, i.e., Ppeak = Tpeak/t, may exceed 200 kPa for thinner tissues, which could result in tissue tearing during testing. In these scenarios, the peak membrane tension was adjusted to a maximum first Piola-Kirchhoff stress of 200 kPa.
- Execute the preconditioning protocol. Following preconditioning, record the current X- and Y-dimensions of the specimen for use in the biaxial testing protocols.
- Create a force-controlled preconditioning protocol, in which the tissue with edge length and thickness will undergo six repetitions of equibiaxial loading to a peak membrane tension Tpeak of 40 N/m with a preload of 3% × Tpeak10 and stretch and recovery times of 30 s each.
- Creation and execution of biaxial testing protocols
- Determine the time required to achieve the peak equibiaxial configuration from the post-preconditioned configuration with the desired displacement rate. Considering a constant displacement rate, compute the loading times for the remaining loading ratios (i.e., TXX:TYY = 1:0.5 and TXX:TYY = 0.5:1).
- Manually jog the linear actuators to match the target forces for a given loading ratio. Repeat this process and record the leaflet dimensions for all loading ratios.
- Prepare a displacement-controlled testing protocol that biaxially displaces the tissue from the post-preconditioned configuration to the configurations recorded in step 1.6.2 (i.e., TXX:TYY = 1:1, 1:0.5, 0.5:1) within the times determined in step 1.6.1. Ensure that each protocol has three loading/unloading cycles for repeatability of the mechanical behavior.
- Construct a testing directory that will store the data for future calculations. Ensure the directory name matches the current specimen.
- Construct a new testing parameter set with the name 1:1, set the X-axis and Y-axis control modes to displacement, and set the control functions to ramp. Define the stretch magnitude to be the configuration recorded in step 1.6.1. Define the preload magnitude as 3% of fpeak for only the first repetition, and define both the stretch duration and recovery duration as the time recorded in step 1.6.1. Define the number of repetitions as 3.
- Repeat step 1.6.3.2 for the remaining loading ratios (i.e., TXX:TYY = 1:0.5 and TXX:TYY = 0.5:1), except define the preload magnitude as not applied. Ensure the stretch magnitude, stretch duration, and recovery duration match those recorded in step 1.6.2.
NOTE: Only data from the final (third) cycle will be used for stress and strain analyses.
- Execute the displacement-controlled protocols. After completion of biaxial testing, return the tissue to its post-preconditioned dimensions.
NOTE: The test should be immediately aborted if the tissue begins to tear.
- Further characterizations
- Leave the tissue submerged in DI water and mounted to the biaxial testing system. Perform pSFDI imaging as described in steps 2.1-2.3.
- Unmount the tissue. If it is an intact tissue, proceed to the microdissection described in steps 3.1-3.7. If not, collect histology following step 3.7.
NOTE: The DI water bath can be used for subsequent characterizations within the same day. - Repeat steps 1.2-1.7 with the A/S and F/V layers acquired following the microdissection (steps 3.1-3.6).
NOTE: The repetition of the testing protocol for the layers allows for direct comparison of the intact tissue to its own layers.
- Biaxial testing data post processing procedures
- Perform digital image correlation of the acquired biaxial testing images to determine the time-dependent marker positions. Compute the fiducial marker displacements via Eq (1).5
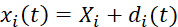 (1)
(1)
Herein, xi(t), Xi, and di(t) are the time-dependent location, the initial (reference) location, and the displacement of marker i. - Compute the deformation gradient F by considering the fiducial markers as a four-node bilinear finite element, as shown in Eq (2)5
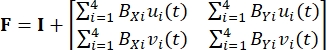 (2)
(2)
Where BXi and BYi are the derivatives of the shape functions for node i in the X- and Y-directions, respectively, and ui(t) and vi(t) are the components of di(t): di(t) = [ui(t), vi(t)]T. - Compute the applied first Piola-Kirchhoff stress P using the recorded forces, as in Eq (3)5
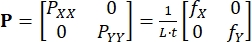 (3)
(3)
PXX and PYY are the X- and Y-components of P; L and t are the tissue edge length and thickness; fX and fY are the forces recorded in the X- and Y-directions. - Determine other strain and stress measures as needed,13 which include the right Cauchy-Green deformation C = FT/F, the Green-Lagrange strain E = (C - I)/2, the Cauchy stress σ = J-1PFT, and the second Piola-Kirchhoff stress S = F-1P.
NOTE: Herein, I is a second-order identity tensor, and J = det(F) is the Jacobian of the deformation gradient F.
- Perform digital image correlation of the acquired biaxial testing images to determine the time-dependent marker positions. Compute the fiducial marker displacements via Eq (1).5
2. Polarized spatial frequency domain imaging
- System preparation
NOTE: If desired, the fiducial markers can be removed from the tissue prior to the following steps.- Center the pSFDI device over the specimen (Figure 2). Turn on the projector and illuminate the specimen with 490 nm (cyan) light.
- Open the camera software and inspect the camera's field of view. Ensure the specimen is centered in the frame and is completely contained within the field of view.
- If the mounted specimen is an intact leaflet, adjust the digital light processing (DLP) projector brightness to ensure the tissue is fully illuminated with no glares on the tissue surface. Do not adjust the brightness if the specimen is one of the composite layers.
- Rotate the polarizer across its complete range of motion to detect possible glares or dirt on the polarizer lens. Carefully clean the polarizer lens with a microfiber cloth as necessary.
- Data collection
NOTE: The following data collection can be automated using software, such as LabVIEW or Python.- Move the polarizer to its home position—ideally aligned with one of the biaxial testing axes. Capture one grayscale image and save it to the computer with the polarizer location (i.e., 0°).
- Rotate the polarizer 5° and capture another grayscale image. Repeat this process to acquire 37 images that range from 0° to 180° with a 5° increment.
NOTE: The images from the first pSFDI imaging sequence can be preliminarily analyzed to ensure the desired optical response from the tissue. Refer to step 2.3 for instructions. - Repeat the pSFDI imaging sequence for other desired tissue configurations, for example, the peak configurations of the loading protocols considered for biaxial mechanical testing.
- pSFDI data postprocessing procedures
NOTE: The following method includes steps for the MATLAB program language. However, any preferred language (e.g., Python, C++) may be used instead.- Use the MATLAB imread() function to construct arrays containing the pixel-wise intensities of the 37 acquired images. For convenience, arrange these as an n × m × 37 three-dimensional array, where n and m are the numbers of pixels along the two axes.
- Define the tissue region of interest (ROI) using the user-defined grabit() function.
- Fit the intensity vs. polarizer angle data for each ROI pixel using a 3-term Fourier series, as in Eq (4):
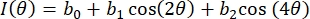 (4)
(4)
Herein, I(θ) is the pixel-wise intensity as a function of polarizer angle, and bi are the Fourier constants. Use standard linear least-squares regression to determine bi. - Determine the pixel-wise fiber orientation as the polarizer angle associated with the maximum value of I(θ). Compute the degree of optical anisotropy (DOA) via Eq (5).
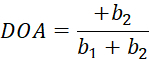 (5)
(5) - Use plot() and histogram() to visualize the acquired fiber orientation and DOA values. Save the processed results for later use.
3. Microdissection of tricuspid valve leaflet composite layers
- Tissue attachment to wax board
- Gather the required materials: wax board, DI water, pipette, scalpel, micro scissors, thin forceps, curved forceps, thick forceps, and pins.
NOTE: Only use tweezers without teeth or grips, as forceps of this type can very easily rip the thin tissue of the A/S layer when performing the dissection. - Unmount the tissue from the biaxial tester and measure its thickness using the laser displacement sensor described in step 1.2. Place the tissue on the wax board.
- Examine the ventricularis side of the tissue for large chordae insertions. Note the position of these insertions to avoid them during the dissection (Supplemental Figure S1). Take a photograph for reference.
- Spread the tissue flat on the wax board with the atrialis facing up. Affix the tissue to the board using the pins:
- In each corner of the tissue, place one pin that is angled away from the tissue (for better viewing) and slightly pulls the tissue taut (Figure 3A). Do this in clockwise or counterclockwise order. Ensure that the pins are outside the holes created by the tines when mounting the tissue.
- Adjust the pin placement slightly to ensure that the tissue is taut and in a square configuration (Figure 3B) so that the tissue lies flat and does not shift during the layer micro-dissection.
- If necessary, place pins along the side of the tissue during the dissection to stretch the tissue more. Keep in mind when placing and angling additional pins that they must be worked around during the dissection.
- Remove the glass bead fiducial markers.
NOTE: The following step is optional. The added DI water helps maintain tissue hydration and prevents the tissue from sticking to itself throughout the microdissection. - Using a pipette, place DI water on the surface of the tissue so that it completely covers the tissue in a bubble-like manner. Replenish the DI water as needed throughout the dissection.
- Gather the required materials: wax board, DI water, pipette, scalpel, micro scissors, thin forceps, curved forceps, thick forceps, and pins.
- Make the initial corner.
- Select a corner of the pinned specimen to begin the dissection. Avoid large chordae insertions and extremely thin areas.
- Make a cut in the A/S layer by lightly dragging the scalpel over the tissue surface along the mounting holes from mechanical testing (Figure 3C). Ensure the cut is at least 5 mm long, and the edges of the cut start to pull apart, revealing the F/V layer underneath.
- Use the thin forceps (without a sharp tip) to firmly rub along the cut and pull the edges of the cut apart (Supplemental Figure S2).
- If the cut in the A/S layer does not start to pull apart, lightly trace over the same cut again with the scalpel until it begins to do this. Be careful not to cut too deeply into the tissue (past the A/S composite layer) as it makes it more difficult to cleanly separate the layers.
- Repeat steps 3.2 and 3.2.3 to make a second cut perpendicular to the first cut (Figure 3D). Ensure that the two cuts are connected and form a corner.
- If the two cuts are not connected, run the thin tweezers under the small area of tissue separating the two cuts (Supplemental Figure S3). Then, carefully use the scissors to cut the tissue.
- Peel the tissue from the corner.
- Rub along the cuts using the thin forceps until the tissue begins to separate from the F/V layer. As soon as a small piece of tissue is separated, grasp it with the tweezers and gently pull it to further separate the composite layers.
NOTE: Always place the tip of the thin tweezers past the edge of the tissue when grasping. Otherwise, they could accidentally poke a hole into the A/S composite layer. - Continue to peel the tissue and rub the seam until it reaches the end of the two cuts made for the corner. Throughout this process, switch to larger tweezers to grasp the tissue for the peeling process to prevent undesired ripping and tearing of the A/S composite layer.
- If the first corner attempted has major issues with separation, try a different corner as a starting point (go back to step 3.2).
- Rub along the cuts using the thin forceps until the tissue begins to separate from the F/V layer. As soon as a small piece of tissue is separated, grasp it with the tweezers and gently pull it to further separate the composite layers.
- Extend cuts, peel the tissue, and make a second corner.
- Extend the two cuts made for the first corner by placing the scalpel tip at the bottom of each cut and lightly dragging it along the tissue surface (Figure 4A). Ensure that all extension cuts are at least 5 mm and the cut extensions connect to the original cuts and continue to follow the tine or suture holes.
NOTE: If the extension cut is too deep, then the forthcoming peeling must be closely monitored to ensure sections of the fibrosa are not separated with the A/S composite layer (Figure 5A). - Continue to extend the cuts and peel the top composite A/S layer back while rubbing the seam until one side is finished. Observe that the tissue will be separated completely along one cut; ensure that the seam between the A/S and F/V composite layers is straight (Figure 4B).
- Repeat the instructions in step 3.2 and step 3.3 to create a second corner perpendicular to the end of the fully peeled side (Figure 4C).
- Extend the two cuts made for the first corner by placing the scalpel tip at the bottom of each cut and lightly dragging it along the tissue surface (Figure 4A). Ensure that all extension cuts are at least 5 mm and the cut extensions connect to the original cuts and continue to follow the tine or suture holes.
- Completely separate the A/S layer.
- Extend the remaining cuts while avoiding large chordae insertions. Continue separating the A/S and F/V layers by using the rubbing and pulling techniques employed for the first corner. Make a note of several considerations or problems that may arise during this process:
- Exclude the chordae insertions from the A/S separation area (Figure 5B) only when this exclusion will allow for an A/S specimen large enough for experimental characterizations (>3.3 mm).
- If the tissue tears or a hole forms, stop separating the tissue immediately. To prevent tweezers from being caught, place the scissors in any hole that forms and cut the tissue away from the center. If the defect forms along the seam of separation, then begin separating the tissue along another edge to prevent further tearing (Figure 5C).
- Look for interlayer connections that may appear while separating the tissue and prevent further separation of the tissue without a high risk of ripping (Figure 5D). Observe these are thin but strong strands that must be carefully cut using scissors. Avoid creating a hole in the A/S layer or cutting downwards into the F/V layer, as this would cause uneven separation.
- Continue this process until the largest sample possible of the A/S layer has been separated. Mark the orientation of the sample using the surgical pen (Figure 6A).
- Extend the remaining cuts while avoiding large chordae insertions. Continue separating the A/S and F/V layers by using the rubbing and pulling techniques employed for the first corner. Make a note of several considerations or problems that may arise during this process:
- Finish dissection.
- Use the scissors to cut along the seam of separation for the remaining tissue edge (Figure 6B). Ensure this cut is as close to the seam of separation as possible.
- Place the separated A/S composite layer flat on the cutting mat. If necessary, use the scalpel to straighten the edges of the tissue and create a square tissue shape suitable for biaxial mechanical testing. Place the A/S layer in DI water until it is ready to be tested.
- Mark the orientation of the F/V layer that remains on the wax board. Cut the largest square possible out of the area where the A/S layer was removed (Figure 6C), then place it in DI water.
- Histology
- Excise two strips of tissue—aligned with the circumferential and radial directions—for use in histology. Use different protocols for the intact and the composite layers (i.e., A/S and F/V).
- For the intact layer, take the specimens from the tissue that remains pinned to the wax board. Use the tissue outside the tine/suture holes, as this portion of the tissue has not been dissected and will represent the intact leaflet.
- For the A/S and F/V composite layers, only collect histology samples after fully completing their testing and imaging. Unmount the specimen from the biaxial testing system, lay the tissue flat on a cutting mat, and excise the circumferential and radial strips using a razor blade.
- Place the excised strips in tissue cassettes and submerge the cassettes in 10% formalin.
- Discard the remaining tissue. Clean the dissection tools using cleaning compound (see the Table of Materials).
- After 24-48 h of fixation, transfer the cassettes to ethanol, where they can be stored indefinitely until histology processing and staining.
NOTE: This histological analysis can confirm that the microdissection is successful. CAUTION: 10% formalin causes skin irritation and serious eye damage. It may also cause an allergic reaction or cancer through inhalation. When handling, wear appropriate personal protection equipment, such as gloves, goggles, and a lab coat, and only use in well-ventilated spaces, such as in a fume hood. When not in use, make sure the storage container is tightly closed.
- Excise two strips of tissue—aligned with the circumferential and radial directions—for use in histology. Use different protocols for the intact and the composite layers (i.e., A/S and F/V).
Access restricted. Please log in or start a trial to view this content.
Results
The microdissection will yield A/S and F/V specimens with relatively uniform thicknesses that can be mounted to a (commercial) biaxial testing device. Histology analysis of the intact leaflet and the two dissected layers will verify if the tissue was correctly separated along the border between the spongiosa and fibrosa (Figure 7). Additionally, the histology micrographs can be used to determine the tissue layer thicknesses and constituent mass fractions using ImageJ software. A failed disse...
Access restricted. Please log in or start a trial to view this content.
Discussion
Critical steps for the protocol include: (i) the layer microdissection, (ii) the tissue mounting, (iii) the fiducial marker placement, and (iv) the pSFDI setup. Appropriate layer microdissection is the most important and difficult aspect of the method described herein. Prior to launching an investigation utilizing this technique, the dissector(s) should have long-term practice with the microdissection technique and all three TV leaflets. The dissector should ensure the composite layer specimens are sufficiently large (&#...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
This work was supported by the American Heart Association Scientist Development Grant (16SDG27760143) and the Presbyterian Health Foundation. KMC was supported in part by the University of Oklahoma (OU) Undergraduate Research Opportunity Program and Honors Research Apprenticeship Program. DWL was supported in part by the National Science Foundation Graduate Research Fellowship (GRF 2019254233) and the American Heart Association/Children's Heart Foundation Predoctoral Fellowship (Award #821298). All of this support is gratefully acknowledged.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 10% Formalin Solution, Neutral Buffered | Sigma-Aldrich | HT501128-4L | |
| Alconox Detergent | Alconox | cleaning compound | |
| BioTester - Biaxial Tester | CellScale Biomaterials Testing | 1.5 N Load Cell Capacity | |
| Cutting Mat | Dahle | B0027RS8DU | |
| Deionized Water | N/A | ||
| Fine-Tipped Tool | HTI INSTRUMENTS | NSPLS-12 | |
| Forceps - Curved | Scientific Labwares | 16122 | |
| Forceps - Thick | Scientific Labwares | 161001078 | |
| Forceps - Thin | Scientific Labwares | 16127 | |
| LabJoy | CellScale Biomaterials Testing | Version 10.66 | |
| Laser Displacement Sensor | Keyence | IL-030 | |
| Liquid Cyanoacrylate Glue | Loctite | 2436365 | |
| MATLAB | MathWorks | Version 2020a | |
| Micro Scissors | HTI Instruments | CAS55C | |
| Pipette | Belmaks | 360758081051Y4 | |
| Polarized Spatial Frequency Domain Imaging Device | N/A | Made in-house using a digital light projector, linear polarizer, rotating polarizer mount, and charge-coupled device camera. See doi.org/10.1016/j.actbio.2019.11.028 (PMCID: PMC8101699) for more details. | |
| Scalpel | THINKPRICE | TP-SCALPEL-3010 | |
| Single Edge Industrial Razor Blades (Surgical Carbon Steel) | VWR International | H3515541105024 | |
| Surgical Pen | LabAider | LAB-Skin-6 | |
| T-Pins | Business Source | BSN32351 | |
| Wax Board | N/A | Made in-house using modeling wax and baking tray | |
| Weigh Boat | Pure Ponta | mdo-azoc-1030 |
References
- Vesely, I. The role of elastin in aortic valve mechanics. Journal of Biomechanics. 31 (2), 115-123 (1998).
- Zhang, W., Ayoub, S., Liao, J., Sacks, M. S. A meso-scale layer-specific structural constitutive model of the mitral heart valve leaflets. Acta Biomaterialia. 32, 238-255 (2016).
- Stella, J. A., Sacks, M. S. On the biaxial mechanical properties of the layers of the aortic valve leaflet. Journal of Biomechanical Engineering. 129 (5), 757-766 (2007).
- Khoiy, K. A., Amini, R. On the biaxial mechanical response of porcine tricuspid valve leaflets. Journal of Biomechanical Engineering. 138 (10), 104504(2016).
- Jett, S. V., et al. An investigation of the anisotropic mechanical properties and anatomical structure of porcine atrioventricular heart valves. Journal of the Mechanical Behavior of Biomedical Materials. 87, 155-171 (2018).
- Meador, W. D., et al. A detailed mechanical and microstructural analysis of ovine tricuspid valve leaflets. Acta Biomaterialia. 102, 100-113 (2020).
- Hudson, L. T., et al. A pilot study on linking tissue mechanics with load-dependent collagen microstructures in porcine tricuspid valve leaflets. Bioengineering. 7 (2), 60(2020).
- Pant, A. D., et al. Pressure-induced microstructural changes in porcine tricuspid valve leaflets. Acta Biomaterialia. 67, 248-258 (2018).
- Kramer, K. E., et al. An investigation of layer-specific tissue biomechanics of porcine atrioventricular heart valve leaflets. Acta Biomaterialia. 96, 368-384 (2019).
- Ross, C. J., Laurence, D. W., Wu, Y., Lee, C. -H. Biaxial mechanical characterizations of atrioventricular heart valves. Journal of Visualized Experiments: JoVE. (146), e59170(2019).
- Goth, W., Lesicko, J., Sacks, M. S., Tunnell, J. W. Optical-based analysis of soft tissue structures. Annual Review of Biomedical Engineering. 18, 357-385 (2016).
- Jett, S. V., et al. Integration of polarized spatial frequency domain imaging (pSFDI) with a biaxial mechanical testing system for quantification of load-dependent collagen architecture in soft collagenous tissues. Acta Biomaterialia. 102, 149-168 (2020).
- Reddy, J. N. An Introduction to Continuum Mechanics. , Cambridge University Press. (2013).
- Duginski, G. A., Ross, C. J., Laurence, D. W., Johns, C. H., Lee, C. -H. An investigation of the effect of freezing storage on the biaxial mechanical properties of excised porcine tricuspid valve anterior leaflets. Journal of the Mechanical Behavior of Biomedical Materials. 101, 103438(2020).
- Salinas, S. D., Clark, M. M., Amini, R. Mechanical response changes in porcine tricuspid valve anterior leaflet under osmotic-induced swelling. Bioengineering. 6 (3), 70(2019).
- Pokutta-Paskaleva, A., Sulejmani, F., DelRocini, M., Sun, W. Comparative mechanical, morphological, and microstructural characterization of porcine mitral and tricuspid leaflets and chordae tendineae. Acta Biomaterialia. 85, 241-252 (2019).
- Ross, C. J., et al. An investigation of the glycosaminoglycan contribution to biaxial mechanical behaviors of porcine atrioventricular heart valve leaflets. Journal of the Royal Society Interface. 16 (156), 0069(2019).
- Sommer, G., Regitnig, P., Költringer, L., Holzapfel, G. A. Biaxial mechanical properties of intact and layer-dissected human carotid arteries at physiological and supraphysiological loadings. American Journal of Physiology-Heart and Circulatory Physiology. 298 (3), 898-912 (2009).
- Holzapfel, G. A., Sommer, G., Gasser, C., Regitnig, P. Determination of the layer-specific mechanical properties ofhuman coronary arteries with intimal thickening, and related constitutive modelling. American Journal of Physiology-Heart and Circulatory Physiology. 289 (5), 2048-2058 (2005).
- Sommer, G., et al. Multiaxial mechanical response and constitutive modeling of esophageal tissues: Impact on esophageal tissue engineering. Acta Biomaterialia. 9 (12), 9379-9391 (2013).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved