A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Combining Human Organoids and Organ-on-a-Chip Technology to Model Intestinal Region-Specific Functionality
In This Article
Summary
Biopsy-derived intestinal organoids and organ-on-a-chips technologies are combined into a microphysiological platform to recapitulate region-specific intestinal functionality.
Abstract
The intestinal mucosa is a complex physical and biochemical barrier that fulfills a myriad of important functions. It enables the transport, absorption, and metabolism of nutrients and xenobiotics while facilitating a symbiotic relationship with microbiota and restricting the invasion of microorganisms. Functional interaction between various cell types and their physical and biochemical environment is vital to establish and maintain intestinal tissue homeostasis. Modeling these complex interactions and integrated intestinal physiology in vitro is a formidable goal with the potential to transform the way new therapeutic targets and drug candidates are discovered and developed.
Organoids and Organ-on-a-Chip technologies have recently been combined to generate human-relevant intestine chips suitable for studying the functional aspects of intestinal physiology and pathophysiology in vitro. Organoids derived from the biopsies of the small (duodenum) and large intestine are seeded into the top compartment of an organ chip and then successfully expand as monolayers while preserving the distinct cellular, molecular, and functional features of each intestinal region. Human intestine tissue-specific microvascular endothelial cells are incorporated in the bottom compartment of the organ chip to recreate the epithelial-endothelial interface. This novel platform facilitates luminal exposure to nutrients, drugs, and microorganisms, enabling studies of intestinal transport, permeability, and host-microbe interactions.
Here, a detailed protocol is provided for the establishment of intestine chips representing the human duodenum (duodenum chip) and colon (colon chip), and their subsequent culture under continuous flow and peristalsis-like deformations. We demonstrate methods for assessing drug metabolism and CYP3A4 induction in duodenum chip using prototypical inducers and substrates. Lastly, we provide a step-by-step procedure for the in vitro modeling of interferon gamma (IFNγ)-mediated barrier disruption (leaky gut syndrome) in a colon chip, including methods for evaluating the alteration of paracellular permeability, changes in cytokine secretion, and transcriptomic profiling of the cells within the chip.
Introduction
The human intestine is a complex and multitasking organ capable of self-regeneration. It is divided into the small and large intestine. The primary function of the small intestine is to further digest food coming from the stomach, absorb all the nutrients, and pass the residue on to the large intestine, which recovers the water and electrolytes. The small intestine is further divided into multiple anatomically distinct regions: the duodenum, jejunum, and ileum, each of which is adapted to perform specific functions. For example, the duodenum helps break down of the chyme (stomach contents) to enable the proper absorption of nutrients involving proteins, carbohydrates, vitamins, and minerals in the jejunum. This proximal part of the small intestine is also the main site of intestinal drug absorption and metabolism, and it is characterized by the higher expression of drug-metabolizing enzymes (e.g., CYP3A4) compared to their expression in the ileum and colon1. In addition to its main role in digesting and absorbing of nutrients, the intestine is also an effective barrier against potentially harmful luminal contents, such as pathogenic microorganisms, microbial metabolites, dietary antigens, and toxins2,3. It is noteworthy that the human colon is inhabited by a large number of microorganisms, far exceeding that of total cells in the human body, which provide many benefits to nutrition, metabolism, and immunity. Therefore, the maintenance of the integrity of the mucosal barrier formed by intestinal epithelial cells is critical for allowing the symbiotic relationship between the gut microbiota and the host cells by physically separating them to avoid unnecessary immune cell activation2. In addition, programmed intestinal cell death plays an essential role as a self-protective mechanism preventing infected cells from persisting or proliferating—thereby disseminating potential pathogens3—while the continuous self-renewal of intestinal epithelium every four to seven days compensates for the cell loss ensuring barrier integrity and tissue homeostasis. Impairments of described intestinal functions, including nutrient absorption, barrier integrity, or imbalance in intestinal cell death and self-renewal, may result in the development of a range of gastrointestinal disorders, including malnutrition and inflammatory bowel disease (IBD)2,3.
Previously, animal models and transformed cancer-derived intestinal cell lines have been used to study the physiological and pathophysiological functions of human intestinal tissue. However, increasingly prominent concerns about the translatability of animal research to humans, caused by the presence of significant disparities between the two species, highlighted a need for human-relevant alternative methods4. The commonly used in vitro intestinal cell lines include T84, Caco-2, and HT29 cells. While they mimic certain aspects of the intestinal barrier function and membrane transport, they are characterized by an altered expression of drug metabolizing enzymes5, surface receptors, and transporters4. In addition, they lack intestinal segment specificity and fail to recapitulate the complexity of intestinal epithelium, with each model containing only one out of the five epithelial cell types present in the intestine6.
Recently, human intestinal organoid cultures established from fresh biopsies of the small intestine and colon7,8 or induced pluripotent stem cells (iPSC)9 were introduced as alternative experimental models with potential to complement, reduce and perhaps replace animal experimentation in the future. While iPSCs can be obtained in a non-invasive manner, establishing organoids from iPSCs requires the use of complex and lengthy protocols (with several experimental steps) and generates cultures resembling human fetal tissue. In contrast, biopsy-derived organoids are highly scalable, as they can leverage the inherent renewal capacity of intestinal tissue and can be indefinitely passaged and propagated in vitro. Importantly, biopsy-derived organoids maintain the disease and intestinal region-specific characteristics of the primary tissue from which they were developed and emulate the cellular diversity of the intestinal epithelium. Organoids can be used as patient-specific avatars in vitro to unravel the biology and pathogenesis of various gastrointestinal disorders and improve their therapeutic management. Although intestinal organoids have achieved an impressive degree of physiological functionality, they still fail to reproduce the complexity of the native organs due to their lack of critical stromal components—including blood vessels, connective tissue, peripheral nerves, and immune cells—as well as mechanical stimulation. Mechanical parameters, such as flow, shear stress, stretch, and pressure, are known to influence tissue morphogenesis and homeostasis in vivo and were previously shown to improve maturation of cells in vitro10,11,12,13. An additional important drawback of organoid systems is the inaccessibility of the lumen and, thus, to the apical side of the epithelium. This presents a challenge for investigating various mechanisms associated with the polarized expression of ion and drug transporters, host-microbiome interactions, and pharmaceutical toxicity testing. Lastly, organoid cultures suffer from considerable variability in size, morphology, and function, owing to the stochastic nature of the in vitro self-organization process and cell fate choices. Therefore, to realize the full potential of intestinal organoids in disease modeling, drug screening, and regenerative medicine, it is necessary to explore new strategies that reduce the variability in organoid development, improve the access to the luminal compartment, and incorporate missing cell-cell interactions.
Organ-on-a-Chip technology has introduced many techniques for the incorporation of mechanical forces and fluid flow to intestinal cell cultures in vitro. However, since most of the initial proof-of-concept studies have used cancer-derived cell lines that did not exhibit sufficient cellular diversity, the relevance of these systems has been questioned. Recently, we have synergistically combined intestinal organoids and organ-on-a-chip technology to incorporate the best features of each approach into one in vitro system14,15,16. The resulting intestine-chip recapitulates the multicellular architecture of intestinal epithelium, the presence of epithelial-endothelial tissue interface, and the mechanical forces of fluid flow and stretch, enabling emulation of organ-level functions in vitro. Additionally, the use of primary-tissue-derived organoids (which can be sampled from different regions of the human intestine) as a starting material increases this model’s versatility, as chips representing human duodenum, jejunum, ileum, and colon can be established following similar seeding and culture procedures. Importantly, Intestine-Chips enable real-time assessment of: the intestinal barrier integrity; activity of the brush border and drug-metabolism enzymes; production of mucins; secretion of cytokines; and interaction of intestinal cells with pathogenic and commensal microorganisms, as demonstrated in the previously published reports. Notably, when intestine chips were established using organoids generated from different individuals’ tissue, these models captured the expected inter-donor variability in the functional responses to various drugs and treatments. Altogether, merging organoids with Organ-on-a-Chip technology opens the door to more advanced, personalized, in vivo-relevant models that could improve the physiological relevance and accuracy of the in vitro findings as well as their extrapolation to humans. Here, a detailed protocol is presented for establishing the intestine chip and its application in the studies of physiological functions of the two intestinal segments: duodenum and colon. Firstly, the methods for the assessing the activity of the drug-metabolizing enzyme CYP3A4 in the duodenum chip, as well as its induction by prototypical compounds such as rifampicin and Vitamin D3, are described. Secondly, the steps required to model “leaky gut” in the colon chip are outlined in the protocol, with the disruption of the epithelial barrier being performed using hallmark cytokines implicated in the pathogenesis of the IBD. Briefly, organoids derived from human biopsies are propagated in vitro, subjected to enzymatic digestion, and introduced in the top channel of the chip. In the presence of continuous perfusion with growth-factor-enriched media they develop into a confluent epithelial monolayer with 3D architecture and a readily accessible apical cell surface. The bottom, "vascular" chip compartment is seeded with microvascular endothelial cells isolated from the small or large intestine. The epithelium and endothelium are separated by a porous stretchable membrane, which facilitates the paracrine interactions between the two tissues and, when subjected to cyclic deformations, emulates peristalsis-like motions of the human gut. The co-culture is maintained under the dynamic flow conditions generated by luminal and vascular perfusion with appropriate cell culture media. Finally, we describe numerous types of assays and endpoint analyses that can be performed directly on-chip or from sampled cell culture effluents.
Protocol
NOTE: All cell cultures should be handled using a proper aseptic technique.
The human intestinal organoids employed in this study were obtained from Johns Hopkins University and all methods were carried out in accordance with approved guidelines and regulations. All experimental protocols were approved by the Johns Hopkins University Institutional Review Board (IRB #NA 00038329).
1. Preparation of the cell culture reagents
- Prepare the human organoid growth medium following the manufacturer's instructions (Table of Materials) and supplement it with 100 µg/mL of primocin.
- Prepare human microvascular endothelial cell growth medium following the manufacturer's instructions (Table of Materials) and supplement with 1:20 vol/vol FBS and 50 μg/ml instead 100 μg/ml of primocin.
- Resuspend Y-27632 (Rho-kinase Inhibitor) in sterile 0.1% BSA in DPBS to prepare a 10 mM stock solution. Aliquot into sterile microtubes and store at -20 °C for up to 6 months.
- Resuspend CHIR99021 (GSK-3 Inhibitor) in sterile dimethylsulfoxide (DMSO) to prepare a 5 mM stock solution. Aliquot into sterile microtubes and store at -20 °C for up to 6 months.
- Prepare the human organoid digestion solution by mixing a 1:1 vol/vol organoids dissociation solution (Table of Materials) with DPBS solution (Table of Materials) and supplementing it with 10 µM stock solution of Y-27632. This reagent must be made fresh for each use.
2. Culture of Human Intestinal Microvascular Endothelial Cells (HIMECs)
- Initiate the HIMEC culture (Table of Materials) 7 days before seeding it onto the chip. Only HIMECs at passage 1-6 can be used for chip seeding.
- Add 5 mL of the attachment factor solution (Table of Materials) to a T150 flask; incubate at 37 °C for 1 min and discard the solution.
- Add 19 mL of endothelial cell growth medium pre-warmed at room temperature (see step 1.2) to the flask.
- Thaw a frozen vial of HIMEC (2 million cells/vial) in a 37°C water bath.
- Transfer the contents of the vial into the flask and gently rock the flask to distribute the cells evenly. Place the flask in a 37 °C incubator overnight to allow the endothelial cells to adhere.
- Replace the medium with 20 mL of endothelial cell growth medium pre-warmed at room temperature on the next day and every 3 days thereafter. Cultures should reach 90% of confluency on the day of cell harvest for chip experiments.
3. Microfabrication and preparation of the chip
- Obtain chips from a commercial supplier (Table of Materials). Unpack the chips and position them in the chip cradle placed in a square Petri dish (Table of Materials). Label the front side of each chip carrier with the experimental conditions (Figure 1A).
NOTE: While the presented protocol relies on the use of a specific commercially available chip and instrumentation, there are several microfluidic devices offered through different vendors, which may offer alternative advantages but lack the ability to incorporate stretch. In addition, microfabrication of the Organs-on-Chips can be performed "in house" following the procedures described in Huh et al.21.
4. Activation and ECM coating of the membrane
- Preparation of the activation solution
- Bring the ER-1 and ER-2 reagents (Table of Materials) to room temperature to equilibrate before use. ER-1 is light sensitive and must be handled in the dark.
- Reconstitute ER-1 in 10 mL of ER-2 reagent to make a final concentration of 0.5 mg/mL and confirm that the ER-1 is completely dissolved.
- Surface activation
- Gently push 50 µL of the ER-1 solution through both channels of the chip (Figure 1A).
- Remove any excess ER-1 solution from the surface of the chip using an aspirator.
- Inspect the chips to make sure that all the chips have received the ER-1 solution. In case of air bubbles, introduce more ER-1 solution until the bubbles are fully removed.
- Incubate the chips inside the UV lamp chamber (Table of Materials) for 15 min. Confirm that the UV lamp is set to the Consistent setting.
- Bring the ER-1 activated chips back to the biosafety cabinet (BSC). Aspirate the ER-1 solution from both the channels. Wash any residue of the ER-1 solution with 200 µL of ER-2.
- Push 200 µL of sterile Dulbecco's phosphate-buffered saline (DBPS) through the channels (Table of Materials) and leave DBPS in the channels until the next step.
- Preparation of extracellular matrix (ECM) and membrane coating
- Preparation of stock solutions of ECM components
- Reconstitute collagen IV (Table of Materials) and fibronectin (Table of Materials) using sterile cell culture grade water (Table of Materials) to a final concentration of 1 mg/mL. Prepare aliquots and store them at -20°C until use.
- Preparation of ECM working solution for membrane coating
- Prepare 1.5 mL of each ECM solution for the top and bottom channel for every 12 chips to be coated. The ECM solution should always be prepared just before use.
- For the top channel, mix collagen IV and solubilized basement membrane matrix (BMM) (Table of Materials) in sterile cold DBPS at 200 µg/mL and 100 µg/mL, respectively. For the bottom channel, mix collagen IV and fibronectin in sterile cold DPBS at 200 µg/mL and 30 µg/mL, respectively.
- ECM coating of the chips
- Aspirate the DBPS from both channels of the chips and replace with the appropriate ECM working solution for each channel (Figure 1A).
- Inspect each chip to ensure it has received the coating solution. In case of air bubbles, push through more coating solution until all the bubbles are fully removed.
- Add sterile DPBS to the chip cradle and place the Petri dish containing the chips in a 37 °C incubator. Incubate overnight to allow the ECM proteins to form ionic bonds with the activated PDMS membrane. If desired, cells can be seeded anytime between 2 h and 1 day after coating of the chips. Alternatively, coated chips can be stored at 4°C overnight, followed by overnight incubation at 37°C and chip seeding.
- Preparation of stock solutions of ECM components
5. Seeding of Intestinal Microvascular Endothelial Cells (HIMECs) in the bottom channel of the chip
NOTE: Small intestinal and colonic HIMECs are seeded in the bottom channel of the duodenum and colon chip, respectively.
- Prepare chips
- Transfer the ECM-coated chips to the BSC. Gently aspirate the ECM coating from both the channels of the chips, and then wash both channels with 200 µL of organoid growth medium and endothelial cell growth medium, respectively.
- Store the washed chips in a 37°C incubator until proceeding with endothelial cells seeding.
- Harvest the endothelial cells
- Bring the HIMEC culture flask to the BSC and wash using sterile DPBS.
- Add 3 mL of dissociation solution to the flask and place it in the 37°C incubator for 2 min to allow for complete cell detachment.
- Collect the cells in a 15 mL conical tube and add endothelial cell growth media until it reaches 10 mL. Sample 15-20 µL of the cell suspension for cell counting. Centrifuge at 150 x g for 5 min.
- Carefully aspirate the supernatant and re-suspend the HIMECs to a density of 8-10 x 106 cells/mL in endothelial cell growth media.
- Seed endothelial cells into the bottom channel of the chip
- Bring the Petri dish containing the chips to the BSC and gently remove all the media from the bottom channel using a 1,000 µL pipette.
- Introduce 10-15 µL of the HIMEC suspension into the bottom channel of the chip. Inspect the chip right after seeding to ensure optimal seeding density (80%-90% of coverage) and homogenous cell distribution within the channel (Figure 1D). If the seeding density is higher or lower than expected or uneven, wash the channel 2x with 200 µL of endothelial cell growth media and repeat the seeding procedure.
- Invert the Petri dish immediately after seeding each batch of six chips to allow endothelial cell attachment to the bottom side of the PDMS membrane. Place the Petri dishes into the 37°C incubator for 30 min to 1 h, or until the HIMECs in the bottom channel have attached to the membrane (Figure 1D).
- Wash the chips
- Gently push 200 µL of endothelial cell growth media through the bottom channel inlet to remove any unattached cells and replenish the nutrients in the media.
- Wash away the cells that did not attach in the top channel with 200 µL of organoid growth medium supplemented with 5 µM CHIR99021 and 10 µM Y-27632.
- Place the chips in the 37°C incubator until proceeding to the epithelial cell seeding.
6. Seeding of organoid fragments in the top channel of the chip
NOTE: Organoids isolated from biopsies of various intestinal regions can be cultured in the intestine chip7. Follow the procedures described in Fujii et al. for the isolation of human intestinal crypts and establishment of organoid cultures22. Here, duodenal and colonic organoids are used to generate the duodenum and colon chips, respectively. Given the high batch-to-batch and donor-to-donor variability in organoid formation and growth, it is suggested to perform a pilot evaluation of cell density in the organoid suspension culture (24-well plate format) to achieve the optimal seeding density of 8 million cells/mL.
- Pilot evaluation of the cell numbers/well of static organoid culture.
NOTE: The following procedure is solely to determine the number of wells of organoid suspension culture required for the chip seeding. The resulting single cells should not be used for the chip seeding.- Carefully aspirate the supernatant from three wells of the organoid culture plate. Add 500 µL of ice-cold BMM dissociation solution (Table of Materials) to each well and use a plastic scraper to detach the solubilized BMM from the plastic.
- Collect the organoid suspension into a sterile low protein binding conical tube using an ice-cold 1,000 µL pipette. Incubate on ice for 60 min, mixing every 15 min by gently shaking the tube. Centrifuge at 300 x g for 5 min at 4°C.
- Aspirate the supernatant and add 2 mL of Trypsin. Incubate at 37°C for 30 min to digest the organoids to the single-cell level and add 10 mL of complete organoid growth medium to stop the enzymatic reaction. Centrifuge at 300 x g for 5 min at 4°C.
- Resuspend the cell pellet in 1 mL of complete organoid growth medium and count the total number of cells using a hematocytometer according to standard methods.
- Preparation of organoid fragments and their seeding in the chip
NOTE: The following procedures can be used to seed duodenal or colonic organoids in the top channel of the chip. The establishment of an epithelial monolayer with an in vivo relevant 3D cytoarchitecture is contingent on the presence of flow15,23 and successful seeding of organoid fragments.- Carefully aspirate the medium from the number of wells of the static organoid culture, which is sufficient, as determined in step 6.1, to achieve final seeding density of 8 million cells/mL. Add 500 µL of ice-cold BMM dissociation solution to each well.
- Detach the solubilized BMM from the surface of the wells using a plastic scraper or 1,000 µL pipette. Collect the suspension into a 15 mL low-protein binding conical tube.
- Store the suspension on ice for 60 min, mixing every 15 min by gently shaking the tube. Proceed to centrifugation at 300 x g for 5 min at 4°C. A well-defined organoid pellet should be visible after the centrifugation. If a transparent gel layer is observed above the cell pellet (residues of solubilized BMM) (Figure 1B), proceed with the following steps.
- Mix the supernatant and cell pellet and add an equal volume of BMM dissociation solution in the tube. Gently mix and store on ice for an additional 10 min, and proceed to centrifugation at 300 x g for 5 min at 4°C.
- If needed, repeat step 6.2.3.1 until no solubilized BMM residues are present.
- Discard the supernatant and resuspend the organoid pellet with organoid digestion solution (see step 1.5). Use a sufficient volume of organoid digestion solution to ensure complete submersion of the pellet. 2 mL of solution is appropriate for approximately 2,400-12,000 medium-sized organoids (Figure 1D, Post-Seeding). Incubate at 37°C for 1-3 min.
- Add Advanced DMEM/F-12 to the tube to stop the enzymatic reaction. Use four times the volume of the digestion solution used. Centrifuge at 300 x g for 5 min at 4°C.
- Aspirate the supernatant and resuspend the organoid fragments pellet in a complete organoid growth medium supplemented with 5 µM CHIR99021 and 10 µM Y-27632 to achieve 8 million cells/mL. Prepare 360 µL aliquots of the suspension in sterile 1.5 mL low protein binding tubes to prevent the reduction of cell density because of attachment on the walls.
- Remove the medium from the top channel of the coated chips. Add 30 µL of cell suspension in the top channel of each chip. Incubate the chips overnight in a 37°C incubator to allow the organoid fragments to adhere to the ECM-coated membrane (Figure 1D).
7. Dynamic culture of intestine chip - initiation, and maintenance of flow and peristalsis-like motions
- Media preparation and degassing
- To maintain constant laminar flow through the chip, allow the media temperature to equilibrate to room temperature, and then subject it to vacuum-driven filtration for 10 min using a vacuum pump and PVDF filter conical tubes (media degassing).
- Priming of portable modules
NOTE: Portable modules are reservoirs that act as an interface between the chips and the culture module allowing for repeating sampling and dosing of media.- Open the portable modules, in the BSC, and place them on the culture module trays, which are specialized containers for the alignment of the portable modules.
- Add 3 mL of pre-equilibrated complete organoid growth medium supplemented with 5 µM CHIR99021 and 10 µM Y-27632 to the top inlet reservoir and 3 mL of pre-equilibrated endothelial cell growth medium in the bottom inlet reservoir (Figure 1C). Add 300 µL of the same media in the respective outlet reservoirs.
- Bring the trays to the 37°C incubator and slide them into the culture module (Figure 1C). Use controls on the screen of the culture module to run the "Prime Cycle" (1 min long). When the status bar indicates "Ready", the Prime Cycle is completed. Repeat the Prime Cycle to ensure sufficient droplets have formed for successfully connecting the chips.
- Ensure that all portable modules are primed and have visible media droplets.
- Introduction of flow
NOTE: Flow is typically initiated 24 h after seeding organoid fragments to allow the intestinal epithelial cells to attach firmly to the membrane.- Wash both channels of the seeded chips with 200 µL of the respective media to remove any bubbles or cell debris. Leave small droplets of media on the ports after the wash. Slide the chip carrier into the portable module (Figure 1C).
- Place the portable modules on the trays, and then into the culture module. Use the controls on the screen of the culture module to program the appropriate organ chip culture conditions (flow rate and stretch).
NOTE: For standard duodenum and colon chip culture conditions, set the flow rate to 30 µL/h15 and 60 µL/h16 respectively, for both the top and bottom channels. However, the flow rates for each channel are controlled independently and can be set between 0-1,000 µL/h. Syringe or peristaltic pumps can be used, instead of culture modules presented here, to introduce laminar flow to microfluidic chips. However, establishment of reliable fluidic connections between chips and pumps using tubing and connectors might be technically challenging when simultaneous perfusion of multiple chips is required. - Start the "Regulate Cycle" (2 h long) that pressurizes the culture media in portable module and chip to prevent nucleation of the air bubbles. Programmed conditions will resume after completion of the Regulate Cycle.
- Media change
- Prepare fresh media for both channels and replenish them every 48 h by adding 2 mL of fresh medium to the inlet reservoirs (Figure 1C).
- Pause the culture modules and bring the trays to BSC. Aspirate media in all the reservoirs and replenish with fresh media. Bring the trays back and restart the flow.
- Introduction of stretch
NOTE: Allow the cells to grow to 100% confluence before application of cyclic strain. Stretch is typically introduced 3 days post-seeding or 48 h after initiation of fluidic culture. 2% cyclic strain at the frequency of 0.2 or 0.15 Hz, for the duodenum11 and colon24 intestine chip, respectively, is applied for the initial 24 h. It is then further increased to 10% for the remaining duration of the intestine chip culture to closely resemble the cyclic strain experienced by intestinal epithelial cells in vivo (Figure 1D)25. The culture module can support application of 2%-12% cyclic strain and frequency of up to 0.4 Hz.- To introduce stretch to the ongoing fluidic culture, pause the culture module. Using controls on the screen, change the stretch settings to 2% stretch, 0.2 or 0.15 Hz frequency, and restart the culture module.
- After 24 h, repeat step 7.5.1 to apply a 10% stretch, 0.2 or 0.15 Hz frequency.
8. CYP450 induction using prototypical CYP inducers in the duodenum chip
NOTE: The cytochrome P450 (CYP450) induction assay enables assessing whether the test compound increases the mRNA levels and/or catalytic activity of specific CYP450 enzymes. Here, we describe the protocol for the evaluation of CYP3A4 induction by the industry standard and regulator recommended in vitro CYP inducers, Rifampicin (RIF) and 1,2-dihydroxy vitamin D3 (VD3). The presented method may be used to identify the potential of various test compounds to induce different isoforms of CYP450 in human intestinal tissue. Specific sets of primers and probe substrate will need to be selected for each enzyme isoform to be evaluated.
- Exposure to CYP Inducers
- Prepare stock solutions of 20 mM RIF, 100 mM VD3, and 200 mM Testosterone (Table of Materials) using sterile DMSO.
CAUTION: Testosterone is a Schedule III controlled substance. Follow regulatory procedures during handling. - Prepare dosing media with CYP inducers by diluting the stock solutions in complete organoid growth medium and endothelial cell growth medium to achieve 20 µM RIF, 100 nmol/L of VD3. Prepare vehicle control by diluting DMSO in the respective media to achieve 0.1% of DMSO.
- Pause the culture module and bring the trays to BSC. Replace the media in all the inlet reservoirs with 2 mL of dosing media with inducers or vehicle control. Return the portable modules back to the culture module and restart the flow at 30 µL/h.
- After 24 h, replace the media with inducing solution, and repeat steps 8.1.2-8.1.3. Inducing solutions should be prepared fresh daily and replenished every 24 h over the course of the experiment, which is typically 48-72 h long.
- Prepare stock solutions of 20 mM RIF, 100 mM VD3, and 200 mM Testosterone (Table of Materials) using sterile DMSO.
- Incubation with a prototypical substrate (Testosterone)
- On the day of harvest, prepare the probe substrate solution of Testosterone along with corresponding inducers in Advanced DMEM/F12 to yield a final concentration of 200 µM. Omit the addition of the serum to the media as this may interfere with the LCMS analysis.
- Bring the trays to BSC, and aspirate the dosing medium from all the reservoirs. Wash and replace the top and bottom inlet reservoirs with warm Advanced DMEM/F12 medium and endothelial cell growth medium, respectively.
- Remove the wash medium from the reservoirs and replace it with 1 mL of probe substrate solution, prepared in step 8.2.1. Perfuse the chips at a high flow rate of 1,000 µL/h for 5 min and aspirate both top and bottom outlet reservoirs. Return the chips to the culture module and incubate for 1 h under the constant flow of 300 µL/h.
- Data analysis
- Sample collection for LCMS analysis
- Aliquot 200 µL of stop solution containing Acetonitrile with 0.1% formic acid in pre-labeled 1.5 mL tubes and place them on ice. The composition of LCMS stop solution may vary based on the substrate that needs to be analyzed.
- After 1 h of treatment is complete, stop the flow and bring the trays back to BSC. Collect 100 µL of effluent from the top outlet reservoir, and add it to the corresponding tube containing the stop solution (Figure 2A). Place the tubes immediately on dry ice. Store the samples at -80°C before proceeding to analysis.
- Extract and analyze samples by using either HPLC or LC-MS/MS techniques, monitoring the formation of metabolite-6β-hydroxytestosterone (6β-OH-T). CYP enzyme activity can be expressed as pmol/min/mg protein where pmol refers to the amount of metabolite (6β-OH-T) formed during the reaction. Total protein content per chip (protein; mg) is determined by performing a Bradford assay described in step 8.3.2.

The fold-induction activity is calculated by: CYP activity(induced) / CYP activity (vehicle). - If collecting samples for mRNA analysis, see step 8.3.3.
- Lysis of the cells for protein expression analysis
NOTE: Protein extraction from cells on-chip is performed using a protein lysis buffer, supplemented with protease and phosphatase inhibitors (Table of Materials). Extracted protein is then quantified using the Bradford assay and used in downstream analysis.- Detach the chips from portable modules and place them in a Petri dish. Wash both the channels with 200 µL of sterile DPBS.
- Block the bottom channel outlet with a 200 µL filter pipette tip. Perfuse 50 µL of the dissociation solution through the bottom channel and incubate for 2 min at room temperature.
- Inspect to confirm complete detachment of the endothelial cells. Pipette up and down 1-2 times and remove the dissociation solution from the channel. Repeat wash.
- Block the top channel outlet with a 200 µL filter pipette tip. Perfuse 75 µL of protein lysis buffer through the top channel. Leave the pipette tip inserted, to block the top channel inlet and incubate for 5 min at room temperature. Pipette up and down 5-10 times and collect the cell lysates in a pre-labeled 1.5 mL tube.
- Repeat step 8.3.2.4. until complete detachment of cells is observed. Collect the cell lysates in the same 1.5 mL tube and store them at -80 °C until analysis.
- Extract the protein fraction according to standard methods and quantify total protein using the Bradford assay (Table of Materials).
- Lysis of the cells for gene expression analysis
NOTE: On-chip cell lysis for RNA extraction can be achieved using an RNA lysis buffer, supplemented with 0.1% 2-mercaptoethanol (Table of Materials). Alternately, a phenol-based RNA lysis buffer can be used if high yields of high-quality RNA suitable for NGS and microarrays analysis are required.- Follow steps 8.3.2.1 to 8.3.2.2 to prepare intestine chip for lysis.
- Block the outlet of the top channel using a 200 µL filter pipette tip. Perfuse 150 µL of RNA lysis buffer through the top channel. Leave the pipette tip inserted to block the inlet of the top channel. Incubate for 2 min at room temperature. For lysis using a phenol-based RNA lysis buffer, use 350 µL of the reagent.
- Pipette up and down 5-10 times and collect the cell lysates in a pre-labelled 1.5 mL tube. Repeat the step with another 150 µL of RNA lysis buffer and collect. Store the cell lysates at -80 °C until analysis. In case of subsequent RNA sequencing analysis, proceed with the analysis within 1-month post lysing.
- Extract total RNA from the cell lysates using an RNA purification kit.
- Reverse-transcribe into cDNA using a reverse transcriptase kit and carry out real-time PCR using a real-time PCR cycler and the appropriate primers and buffer. Quantify the results using the 2-ΔΔCt method.
- Sample collection for LCMS analysis
9. Disruption of the epithelial barrier using proinflammatory cytokines in colon chip
NOTE: This protocol describes disruption of the intestinal epithelial barrier by the cytokine interferon gamma (IFNγ)26,27,28,29. The cytokine is dosed in the bottom channel of the colon chip given the basolateral expression of IFNγ receptor on intestinal epithelial cells. The proinflammatory stimulus is introduced in the chip on day 5 of the culture, as soon as the Papp has been stabilized below 0.5 x 10-6 cm/s. A similar dosing regimen can be used for other proinflammatory cytokines and barrier disruptive agents.
- Stimulation with IFNγ
- Prepare a 100 µg/mL of stock solution of IFNγ (Table of Materials) in sterile cell culture water. Store the stock solution at -80°C over the course of the experiment. For each experiment, always use a fresh stock of IFNγ and avoid more than three freeze and thaw cycles.
- Prepare IFNγ dosing solution by diluting stock solution in degassed endothelial cell growth medium to achieve a final concentration of 10-100 ng/mL.
- Bring the trays to BSC and remove the medium from bottom channel inlet reservoirs and replace it with 3 mL of the IFNγ containing medium. Refresh the IFNγ dosing medium daily.
- Place the trays into the culture module and perfuse the chips at a high flow rate of 1,000 µL/h for 5 min to initiate IFNγ treatment. Switch the flow rate back to 60 µL/h and continue the fluidic culture.
- Data analysis
- Assessment of the epithelial barrier function. The epithelial apparent permeability (Papp), or barrier function, can be measured at various time points of the culture post-treatment with IFNγ, in the effluent media of both channels' inlet and outlet reservoirs. The fluorescent tracer should be added to the top channel organoid growth medium 4 h before the assessment of the Papp. Typically, 3 kDa Dextran Cascade Blue is used as the fluorescent tracer and is added in the medium 24 h before.
- Pause the culture module and bring the trays to the BSC.
- Label and prepare a 96-well black-walled plate with 100 µL of DPBS per well. Using a 200 µL multi-channel pipette, collect and add 50 µL of effluent from all the reservoirs to the respective wells (Figure 2B).
- To prepare the standard curve, dilute the medium containing 100 µg/mL of 3 kDa dextran Cascade Blue 1:3 in DPBS. Subsequently perform serial dilutions using a three-fold dilution of endothelial cell growth medium in DPBS.
- Read the fluorescence at 375 nm excitation and 420 nm emission using a plate reader. Use the OD values measured to calculate apparent permeability (Papp) as follows:
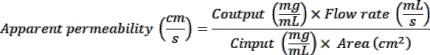
- Immunofluorescence staining of cells on-chip
- Wash using 200 µL of DPBS for each channel, three times.
- Perfuse 200 µL of the fixative solution (4% PFA in DPBS) in each channel. Incubate for 15 min at room temperature.
- Repeat the washing step 9.2.2.1. At this stage, washed chips can be stored for up to 7 days at 4°C.
- Permeabilize the cells using 200 µL of permeabilization solution (0.1% Triton-X 100 in 10% normal donkey serum (NDS) in DPBS) for each channel. Incubate for 30 min at room temperature. Repeat the washing step 9.2.2.1.
- Block the cells on the chip by using 200 µL of blocking solution (10% NDS in DPBS) for each channel. Incubate for 1 h at room temperature.
- Dilute the primary antibodies in antibody solution (5% NDS in DPBS) as follows: anti-Zonula Occludens 1 (ZO-1) (1:100, epithelial tight junction marker), anti-Occludin (1:100, epithelial tight junction marker), anti-Claudin 4 (1:100, epithelial tight junction marker), anti-E-cadherin (1:100, epithelial adherens junctions marker (Table of Materials). Perfuse 200 µL of the primary antibody solution through each channel and incubate overnight at 4 °C. Repeat the washing step 9.2.2.1.
- Dilute the secondary antibodies in antibody solution (1:300, 5% NDS in DPBS). Perfuse 200 µL of this solution through each channel and incubate for 2 h at room temperature. Repeat the washing step 9.2.2.1. If desired, phalloidin, a cytoskeletal marker, can be added to the secondary antibody solution.
- Prepare a 50 µg/mL of 4′,6-diamidino-2-phenylindole (DAPI) solution in DPBS and use it to perfuse 200 µL through each channel. Incubate at room temperature for 15 min. Repeat the washing step. At this point, stained chips can be stored for up to 14 days at 4°C.
- Assessment of Caspase 3 cleavage
- Isolate and quantify the total amount of protein of epithelial cells as described in step 8.3.2. Omit the washing step 8.3.2.1. Dilute the samples with DPBS to a final concentration of 400 µg/mL.
- Quantify the levels of total and cleaved caspase 3 using caspase 3 detection kit following the manufacturer's protocol.
- Assessment of proinflammatory cytokines secretion
- Use the effluents collected from the outlet of both channels of the colon chip to quantify secreted acute-phase proinflammatory cytokines, following protocols provided by the manufacturer. Perform a 5-fold dilution for V-PLEX Vascular Injury Panel 2 Human Kit and a two-fold dilution for V-PLEX Human Proinflammatory Panel II.
- Assessment of the epithelial barrier function. The epithelial apparent permeability (Papp), or barrier function, can be measured at various time points of the culture post-treatment with IFNγ, in the effluent media of both channels' inlet and outlet reservoirs. The fluorescent tracer should be added to the top channel organoid growth medium 4 h before the assessment of the Papp. Typically, 3 kDa Dextran Cascade Blue is used as the fluorescent tracer and is added in the medium 24 h before.
Results
Figure 1D summarizes the timeline of the intestine chip culture and illustrates the intestinal endothelial cells and organoids before and upon seeding on the chip. Moreover, it demonstrates the distinct morphological differences between the duodenum and colon chips, highlighted by the presence of the villi-like formations in the duodenum chip and representative of the small intestinal architecture.
Figure 3A,B demonst...
Discussion
The combination of organ-on-a-chip technology and intestinal organoids holds promise for accurate modeling of human intestinal physiology and pathophysiology. Here, we provide a simple and robust step-by-step protocol (outlined in Figure 1) for establishment of the intestine chip containing biopsy-derived small intestinal or colonic epithelium and intestinal microvascular endothelial cells co-cultured in a microfluidic device. This chip-based simulation of the human intestine incorporates ph...
Disclosures
Gauri Kulkarni, Athanasia Apostolou, Lorna Ewart, Carolina Lucchesi, and Magdalena Kasendra are current or former employees of Emulate Inc. and may hold equity. Emulate Inc. is the company that manufactures the organ chip devices and has published patents relevant to the work stated in this article.
Acknowledgements
We thank Professor Mark Donowitz for providing the intestinal biopsy-derived organoids and Brett Clair for designing the scientific illustrations of the chip, portable and culture module. All the rest of the scientific illustrations were generated using the BioRender.
Materials
| Name | Company | Catalog Number | Comments |
| small intestine Human Intestinal Microvascular Endothelial Cells | AlphaBioRegen | ALHE15 | 0.5 cells M/ml ; cryopreserved |
| colon Human Intestinal Microvascular Endothelial Cells | AlphaBioRegen | ALHE16 | 0.5 cells M/ml ; cryopreserved |
| Biopsy-derived Human Duodenal Organoids | John Hopkin's University | - | The organoids were provided by Professor Mark Donowitz (Institutional Review Board Number: NA_00038329). |
| Biopsy-derived Human Colonic Organoids | John Hopkin's University | - | The organoids were provided by Professor Mark Donowitz (Institutional Review Board Number: NA_00038329). |
| Zoë CM-1™ Culture Module | Emulate Inc. | - | Culture module |
| Orb-HM1™ Hub Module | Emulate Inc. | - | 5% CO2, vacuum stretch, and power supply |
| Chip-S1™ Stretchable Chip | Emulate Inc. | - | Organ-Chip |
| Pod™ Portable Modules | Emulate Inc. | - | Portable module |
| UV Light Box | Emulate Inc. | - | - |
| Chip Cradle | Emulate Inc. | - | 1 per square culture dish |
| Steriflip®-HV Filters | EMD Millipore | SE1M003M00 | 0.45 μm PVDF filter |
| Square Cell Culture Dish (120 x 120 mm) | VWR | 82051-068 | - |
| Handheld vacuum aspirator | Corning | 4930 | - |
| Aspirating pipettes | Corning / Falcon | 357558 | 2 mL, polystyrene, individually wrapped |
| Aspirating tips | - | Sterile (autoclaved) | |
| Serological pipettes | - | 2 mL, 5 mL, 10 mL, and 25 mL low endotoxin, sterile | |
| Pipette | P20, P200, P1000 and standard multichannel | ||
| Pipette tips | P20, P200, and P1000. | ||
| Conical tubes (Protein LoBind® Tubes) | Eppendorf | 0030122216; 0030122240 | 15 mL, 50 mL tubes |
| Eppendorf Tubes® lo-bind | Eppendorf | 022431081 | 1.5 mL tubes |
| 96 wells black walled plate | - | - | for epithelial permeability analysis |
| Microscope (with camera) | - | - | For bright-field imaging |
| Water bath (or beads) | - | - | Set to 37°C |
| Vacuum set-up | - | - | Minimum pressure: -70 kPa |
| Cell scrapers | Biotium | 220033 | |
| T75 flasks | BD Falcon | 353136 | Cell culture flask |
| Emulate Reagent-1 (ER-1) | Emulate Inc. | - | Chip coating solution |
| Emulate Reagent-2 (ER-2) | Emulate Inc. | - | Chip coating solution |
| Dulbecco’s PBS (DPBS) | Corning | 21-031-CV | 1X |
| Cell Culture Grade Water | Corning | MT25055CV | |
| Trypan blue | Sigma | 93595 | For cell counting |
| TryplE Express | ThermoFisher Scientific | 12604013 | Organoids dissociation and endothelium cells detachment solution |
| Advanced DMEM/F12 | ThermoFisher Scientific | 12634028 | Medium |
| IntestiCult™ Organoid Growth Medium (Human) | Stem Cell technologies | 06010 | Organoid Growth Medium |
| Endothelial Cell Growth Medium MV 2 | Promocell | C-22121 | Endothelial medium |
| Fetal bovine serum (FBS) | Sigma | F4135 | Serum |
| Primocin™ | InvivoGen | ANT-PM-1 | antimicrobial agent |
| Attachment Factor™ | Cell Systems | 4Z0-210 | coating solution for flask |
| Matrigel - Growth Factor Reduced | Corning | 356231 | Solubilized basement membrane matrix |
| Collagen IV | Sigma | C5533 | ECM component |
| Fibronectin | Corning | 356008 | ECM component |
| Y-27632 | Stem Cell technologies | 72304 | organoid media supplement |
| CHIR99021 | Reprocell | 04-0004-10 | organoid media supplement |
| Cell Recovery Solution | Corning | 354253 | Basement mebrane matrix dissociationsolution |
| Bovine Serum Albumin (BSA) | Sigma | A9576 | 30%, Sterile |
| Cell Culture Grade Water | Corning | MT25055CV | Sterile, Water |
| DMSO | Sigma | D2650 | solvent |
| 3KDa Dextran Cascade Blue | Invitrogen | D7132 | 10 mg powder |
| Rifampicin (RIF) | Sigma | Cat# R3501 | CYP inducer |
| Testosterone hydrate | Sigma | T1500 | CYP substrate |
| 1,25-dihyroxy Vitamin D3 (VD3) | Sigma | Cat# D1530 | CYP inducer |
| Acetonitrile with 0.1% (v/v) Formic acid | Sigma | 159002 | LCMS stop solution |
| IFNγ | Peprotech | 300-02 | |
| 4% Paraformaldehyde (PFA) | EMS | 157-4 | Fixative |
| Triton-X 100 | Sigma | T8787 | |
| Normal Donkey Serum (NDS) | Sigma | 566460 | |
| anti-Occludin | ThermoFisher Scientific | 33-1500 | tight junctions marker |
| anti-Claudin 4 | ThermoFisher Scientific | 36-4800 | tight junctions marker |
| anti-E-cadherin | Abcam | ab1416 | epithelial adherens junctions marker |
| anti-VE-cadherin | Abcam | ab33168 | endothelial adherent junctions marker |
| anti- Zonula Occludens 1 (ZO-1) | Thermo Fischer | 339194 | tight junctions marker |
| DAPI | ThermoFisher Scientific | 62248 | nuclear stain |
| 2-mercaptoethanol | Sigma | M6250 | |
| PureLink RNA Mini Kit | Invitrogen | 12183020 | RNA lysis, isolation and purification kit |
| SuperScript™ IV VILO™ Master Mix | Invitrogen | 11756050 | reverse transcriptase kit |
| TaqMan™ Fast Advanced Master Mix | Applied Biosystems | 4444557 | qPCR reagent |
| QuantStudio™ 5 Real-Time PCR System | Applied Biosystems | A28573 | Real-time PCR cycler |
| 18S primer | ThermoFisher Scientific | Hs99999901_s1 | Eukaryotic 18S rRNA |
| CYP3A4 primer | ThermoFisher Scientific | Hs00604506_m1 | Cytochrome family 3 subfamily A member 4 |
| Pierce™ Coomassie Plus (Bradford) Assay Kit | ThermoFisher Scientific | 23236 | Protein quantification kit |
| MSD Tris lysis buffer | Meso Scale Diagnostics | R60TX-3 | Protein lysis buffer |
| Cleaved/Total Caspase-3 Whole Cell Lysate Kit | Meso Scale Diagnostics | K15140D | Caspase 3 detection kit |
| V-PLEX Vascular Injury Panel 2 Human Kit | Meso Scale Diagnostics | K15198D | |
| V-PLEX Human Proinflammatory Panel II (4-Plex) | Meso Scale Diagnostics | K15053D | |
| Zeiss LSM 880 | Zeiss | - | Confocal microscope |
| Zeiss LD plan-Neofluar 20x/0.40 Korr M27 | Zeiss | - | 20X long-distance objective lenses |
| Zeiss AXIOvert.A1 | Zeiss | - | Brightfield microscope |
| Zeiss LD A-Plan 10X/0.25 Ph1 | Zeiss | - | 10X objective lenses |
References
- Fritz, A., et al. Expression of clinically relevant drug-metabolizing enzymes along the human intestine and their correlation to drug transporters and nuclear receptors: An intra-subject analysis. Basic and Clinical Pharmacology and Toxicology. 124 (3), 245-255 (2019).
- Okumura, R., Takeda, K. Maintenance of intestinal homeostasis by mucosal barriers. Inflammation and Regeneration. 38 (1), 1-8 (2018).
- Delgado, M. E., Grabinger, T., Brunner, T. Cell death at the intestinal epithelial front line. FEBS Journal. 283 (14), 2701-2719 (2016).
- Mestas, J., Hughes, C. C. W. Of mice and not men: Differences between mouse and human immunology. The Journal of Immunology. 172 (5), 2731-2738 (2004).
- Sun, H., Chow, E. C. Y., Liu, S., Du, Y., Pang, K. S. The Caco-2 cell monolayer: Usefulness and limitations. Expert Opinion on Drug Metabolism and Toxicology. 4 (4), 395-411 (2008).
- Yu, H., et al. The contributions of human mini-intestines to the study of intestinal physiology and pathophysiology. Annual Review of Physiology. 79, 291-312 (2017).
- Sato, T., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459 (7244), 262-265 (2009).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141 (5), 1762-1772 (2011).
- Spence, J. R., et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 470 (7332), 105-110 (2011).
- Durel, J. F., Nerurkar, N. L. Mechanobiology of vertebrate gut morphogenesis. Current Opinion in Genetics and Development. 63, 45-52 (2020).
- Gayer, C. P., Basson, M. D. The effects of mechanical forces on intestinal physiology and pathology. Cellular Signalling. 21 (8), 1237-1244 (2009).
- Xu, Y., et al. Mechanical stimulation activates Piezo1 to promote mucin2 expression in goblet cells. Journal of Gastroenterology and Hepatology (Australia). 36 (11), 3127-3139 (2021).
- Navabi, N., McGuckin, M. A., Lindén, S. K. Gastrointestinal cell lines form polarized epithelia with an adherent mucus layer when cultured in semi-wet interfaces with mechanical stimulation. PLoS One. 8 (7), 68761 (2013).
- Kasendra, M., et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Scientific Reports. 8 (1), 1-14 (2018).
- Kasendra, M., et al. Duodenum intestine-chip for preclinical drug assessment in a human relevant model. eLife. 9, 50135 (2020).
- Apostolou, A., et al. A novel microphysiological colon platform to decipher mechanisms driving human intestinal permeability. Cellular and Molecular Gastroenterology and Hepatology. 12 (5), 1719-1741 (2021).
- Jalili-Firoozinezhad, S., et al. Author correction: A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nature Biomedical Engineering. 3 (7), 583 (2019).
- Yin, J., et al. Fluid shear stress enhances differentiation of jejunal human enteroids in Intestine-Chip. American Journal of Physiology-Gastrointestinal and Liver Physiology. 320 (3), 258-271 (2021).
- Sontheimer-Phelps, A., et al. Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cellular and Molecular Gastroenterology and Hepatology. 9 (3), 507-526 (2020).
- Tovaglieri, A., et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome. 7 (1), 43 (2019).
- Huh, D., et al. Microfabrication of human organs-on-chips. Nature Protocols. 8 (11), 2135-2157 (2013).
- Fujii, M., Matano, M., Nanki, K., Sato, T. Efficient genetic engineering of human intestinal organoids using electroporation. Nature Protocols. 10 (10), 1474-1485 (2015).
- Kim, H. J., Huh, D., Hamilton, G., Ingber, D. E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab on a Chip. 12 (12), 2165-2174 (2012).
- Sarna, S. K. Colonic motility: From bench side to bedside. Morgan & Claypool Life Sciences. , (2010).
- Basson, M. D. Paradigms for mechanical signal transduction in the intestinal epithelium - category: Molecular, cell, and developmental biology. Digestion. 68 (4), 217-225 (2003).
- Wang, F., et al. Interferon-gamma and tumor necrosis factor-alpha synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression. The American Journal of Pathology. 166 (2), 409-419 (2005).
- Nava, P., et al. Interferon-γ regulates intestinal epithelial homeostasis through converging β-Catenin signaling pathways. Immunity. 32 (3), 392-402 (2010).
- Madara, J. L., Stafford, J. Interferon-γ directly affects barrier function of cultured intestinal epithelial monolayers. Journal of Clinical Investigation. 83 (2), 724-727 (1989).
- Bruewer, M., et al. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. The Journal of Immunology. 171 (11), 6164-6172 (2003).
- Jones, S. C., et al. Adhesion molecules in inflammatory bowel disease. Gut. 36 (5), 724-730 (1995).
- Uhlar, C. M., Whitehead, A. S. Serum amyloid A, the major vertebrate acute-phase reactant. European Journal of Biochemistry. 265 (2), 501-523 (1999).
- Pérez-González, C., Ceada, G., Matejčić, M., Trepat, X. Digesting the mechanobiology of the intestinal epithelium. Current Opinion in Genetics & Development. 72, 82-90 (2022).
- In, J., et al. Enterohemorrhagic escherichia coli reduces mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cellular and Molecular Gastroenterology and Hepatology. 2 (1), 48-62 (2016).
- Grassart, A., et al. Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting shigella infection. Cell Host and Microbe. 26 (3), 435-444 (2019).
- Kerns, S. J., et al. Human immunocompetent organ-on-chip platforms allow safety profiling of tumor-targeted t-cell bispecific antibodies. eLife. 10, 67106 (2021).
- Bhatia, S. N., Ingber, D. E. Microfluidic organs-on-chips. Nature Biotechnology. 32 (8), 760-772 (2014).
- Ingber, D. E. Reverse engineering human pathophysiology with organs-on-chips. Cell. 164 (6), 1105-1109 (2016).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved