A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Determination of the Mating Efficiency of Haploids in Saccharomyces cerevisiae
In This Article
Summary
In this work, a robust method for the quantification of mating efficiency in the yeast Saccharomyces cerevisiae is described. This method is particularly useful for the quantification of pre-zygotic barriers in speciation studies.
Abstract
Saccharomyces cerevisiae is a widely used model organism in genetics, evolution, and molecular biology. In recent years, it has also become a popular model organism to study problems related to speciation. The life cycle of yeast involves both asexual and sexual reproductive phases. The ease of performing evolution experiments and the short generation time of the organism allow for the study of the evolution of reproductive barriers. The efficiency with which the two mating types (a and α) mate to form the a/α diploid is referred to as the mating efficiency. Any decrease in the mating efficiency between haploids indicates a pre-zygotic barrier. Thus, to quantify the extent of reproductive isolation between two haploids, a robust method to quantify the mating efficiency is required. To this end, a simple and highly reproducible protocol is presented here. The protocol involves four main steps, which include patching the haploids on a YPD plate, mixing the haploids in equal numbers, diluting and plating for single colonies, and finally, calculating the efficiency based on the number of colonies on a drop-out plate. Auxotrophic markers are employed to clearly make the distinction between haploids and diploids.
Introduction
Saccharomyces cerevisiae, commonly called budding yeast, is a unicellular eukaryote. It has two mating types, a and α, and exhibits both asexual and sexual reproductive cycles. The a and α mating types are haploids and can divide mitotically in the absence of the other mating type in the surrounding environment, which represents the asexual cycle of yeast. When the two mating types are in close proximity, they stop dividing mitotically and fuse to form a diploid cell. The diploid yeast can either divide mitotically when nutrients are present or undergo meiosis under the conditions of nitrogen starvation in the presence of a poor carbon source that is non-fermentable, such as acetate1. This results in the formation of spores, which remain dormant until there are favorable growth conditions. The life cycle is completed when these spores germinate and the two haploid types are released back to the haploid pool2,3 (Figure 1).
The mating of yeast cells includes several steps, such as agglutination, the formation of a mating projection or “shmoo”, followed by cell and nuclear fusion4,5. The two mating types a and α produce a-factor and α-factor, respectively, to initiate mating. These factors are polypeptide pheromones that bind to the receptors (Ste2 and Ste3) present on the cell surface of the opposite mating type5. The binding of the pheromones to the receptors initiates the pheromone response pathway, the mitogen-activated protein kinase (MAPK) signal transduction pathway6,7,8. This results in the arrest of the cell cycle in the G1 phase, leading to a metabolically active stationary phase9. The cells then stop dividing mitotically, and the proteins required for mating are synthesized. As the haploid cells cannot move toward each other, a mating projection or “shmoo” is directed toward the mating partner. When the cells come into contact, the cell wall is degraded, and the cytoplasmic contents fuse, resulting in mating to form a diploid cell10,11. The mating efficiency between haploids has been used as a measure of speciation in laboratory-evolved strains, as well as between extant species12.
Being a simple eukaryotic organism, yeast is the model of choice for a large number of research questions associated with complex eukaryotic organisms. One such question is associated with speciation and the evolution of reproductive barriers13,14. For sexually reproducing organisms, a species is defined by the biological species concept (BSC) proposed by Ernst Mayr15. According to this concept, two individuals of a population are said to belong to two different species if they cannot interbreed and are reproductively isolated. Breakdown of the sexual reproductive cycle (which involves the fusion of gametes to form a zygote, the development of the zygote into a progeny, and the attainment of sexual maturity in the progeny) leads to reproductive isolation. As shown in Figure 1, the life cycle of S. cerevisiae is comparable to the sexual reproductive cycle: a) the fusion of the two mating types a and α is similar to the fusion of gametes in sexually reproducing organisms; b) the ability of the diploid to undergo mitotic division is equivalent to the zygote developing into progeny; and c) the diploid undergoing sporulation is comparable to the process of gametogenesis14.
Pre-zygotic isolation occurs when assortative mating is observed. Given an equal opportunity to mate with two genetically different a types, an α type preferentially mates with one over the other or vice versa14. In the case of evolution experiments in which haploids have been evolved in different environments, the presence of a pre-mating barrier can be determined by performing a mating assay. A decrease in the mating efficiency when compared to the ancestor indicates the evolution of a pre-mating barrier. Post-zygotic isolation could arise due to the inability of the diploid to undergo effective mitotic division and/or sporulation to form haploid spores14. These can be quantified by measuring the growth rate of the diploids and calculating the sporulation efficiency, respectively. Hence, to study the evolution of reproductive barriers, robust methods for quantifying (a) the mating efficiency, (b) the mitotic growth of the diploid, and (c) the sporulation efficiency of the diploid are required. In this work, a robust method to quantify the mating efficiency of yeast strains is reported.
In laboratory experiments, one of the ways in which the occurrence of mating can be detected is by using auxotrophic markers that complement the nutritional requirements. When the two mating types are auxotrophic for two different amino acids, only the diploid cell formed by the fusion of the two mating types can grow on a medium deficient in both the amino acids. Thus, auxotrophic markers are useful for detecting mating both qualitatively and quantitatively. A qualitative test will suffice to identify the mating type of a strain after meiosis16. Quantitative tests are essential when one is interested in identifying a reduction in mating while studying the genes involved in the mating pathway17,18. In addition, with yeast being increasingly used in speciation studies, a convenient and reproducible mating assay is necessary, as the quantification of mating efficiency is a measure of the pre-zygotic barrier.
The mating efficiency between the two yeast mating types has been quantified previously16,19,20. Most of the previously used methods are similar in their design with a few variations16,21,22,23,24,25. Some of them use early log phase cultures, while a few others use mid-log phase cultures of haploid strains. There are variations in the ratios in which the two mating types are mixed. Almost all the protocols use a nitrocellulose membrane. Suspensions of both the mating types taken from previously grown cultures are mixed and filtered onto a nitrocellulose membrane placed on a YPD plate. In one of the variations of the protocol, the haploid suspension is directly patched on a YPD plate21. In experiments dealing with the genes involved in the pheromone production of the two mating types, the pheromones are externally added while making the suspensions of the two mating types24.
Following incubation for a few hours (typically around 5 h) after mixing the haploids, the cells are washed off from the membrane, diluted, and plated on selective media. In one of the earlier methods reported in 1973, the efficiency of zygote formation or mating was calculated by counting the number of budded cells, unbudded cells, and mating pairs under a microscope using a hemocytometer26. However, most methods reported later use auxotrophic markers to distinguish haploids and diploids. Mating efficiency is calculated as the percentage of diploid cells relative to the number of diploid and haploid cells in the cellular pool16,21,23.
However, despite a number of reports using yeast as a model organism to study speciation, there is no standardized protocol reported in the literature until now for calculating the efficiency of mating. Cells in the log phase may not be ideal for the quantification of mating efficiency. During mating, the cell cycle of the two haploids is arrested, and hence, the cells during mating are not dividing9. As the cell cycle is also known to be similarly arrested in cells in the stationary phase27, using such cells can make the protocol more reproducible. Stationary phase cells can be mixed and laid out on YPD plates (i.e., a nutritionally rich environment) for mating. The conventional procedures also require a nitrocellulose membrane and washing off the cells, making the process cumbersome and liable to handling errors. In addition, the protocols used to date quantify the mating efficiency in terms of one haploid. However, when measuring reproductive isolation, mating efficiency is quantified for a particular combination of haploids rather than a single haploid.
To address these issues, here, we report a robust method for the quantification of mating efficiency in yeast that is highly reproducible and easy to use. Moreover, this method and the yeast strains employed here can also be used in studies examining the effect of gene flow on the evolution of mating barriers.
Two different strains of S. cerevisiae were used in this study. One of the strains is derived from the SK1 background; this was modified in our laboratory by adding the auxotrophic markers near the MAT locus. The resulting genotypes of the haploids are provided in Table 128,29,30. In the SK1 strain, the a haploid had the TRP1 gene inserted near the MAT locus, and the α haploid had the LEU2 gene inserted near the MAT locus. In the ScAM strain, the TRP1 and URA3 genes were inserted in the a and α haploids, respectively. The location of insertion was in the ARS region of chromosome III (Chr III: 197378..197609). For the protocol reported here, auxotrophic markers anywhere on the genome would suffice. However, having the auxotrophic markers near the MAT locus means that these strains can also be used for studies examining the effect of gene flow on speciation31,32. The markers were added close to the MAT locus to prevent the reshuffling of the markers due to recombination. Hence, this protocol can be used to quantify mating efficiency in studies involving speciation and also to identify the alteration of mating efficiency when studying the proteins involved in the mating pathway.
Protocol
NOTE: The protocol broadly involves the following steps: (1) patching the haploids in the mating efficiency grids on a YPD plate, (2) mixing the haploids in equal numbers after 24 h incubation and giving the mixed haploids a few hours to mate (7 h in this study), (3) plating the mixed cells on YPD for isolating single colonies after 7 h at 30 °C, and finally, (4) determining the number of diploids formed using the auxotrophic markers. These steps are discussed in detail below (also see Figure 2).
1. Patching of haploids in the mating efficiency grids
- Revive the haploids a and α from freezer stocks by streaking on a YPD agar plate (2% agar, 2% dextrose, 1% peptone, 0.5% yeast extract), and allow them to grow for 48 h at 30 °C to obtain isolated single colonies.
- Inoculate single colonies from the YPD plate in 5 mL of YPD medium (2% dextrose, 1% peptone, 0.5% yeast extract), and incubate at 30 °C for 48 h with 250 rpm shaking. The cells are in the stationary phase of growth after this incubation period.
- Draw a mating efficiency grid on a fresh YPD plate. Draw the grid as a 1 cm x 1.5 cm rectangle divided into three boxes such that each has a dimension of 1 cm x 0.5 cm, as shown in Figure 2A.
- Patch 5 µL of the YPD culture of the two mating types on the leftmost and rightmost rectangles (Figure 2B). This volume corresponds to roughly 5 x 105 haploid cells laid out in each section. Incubate the plates for 24 h at 30 °C.
NOTE: The purpose of the grid is to make the experimental measures precise (such as the number of cells in the experiment). The grid size is small enough that it is experimentally tractable but large enough that it can be easily manipulated (like lifting cells from a grid) and not susceptible to drift or chance events.
2. Mixing of haploids and mating
NOTE: After 24 h (Figure 2C), an equal number of cells of the two haploid types are scraped off the two grids, mixed, and laid in the center rectangle (Figure 2D).
- In order to mix an equal number of cells, remove about 1/3 of the patch that was laid in the outer boxes using a sterile toothpick, and resuspend in 20 µL of water in a sterile 1.5 mL vial for each of the haploids.
- Dilute 5 µL of this suspension in 2 mL of water. Measure the OD of this diluted suspension using a spectrophotometer at 600 nm. Mix an equal number of cells from the two strains, based on the OD value and number of cells/mL in 1 OD for that particular strain. Calculate the volume required to be mixed, and aspirate it from the remaining 15 µL of the individual haploid suspension.
NOTE: The number of cells patched in the center rectangle is such that the cells form a monolayer. Considering the yeast cell to be a sphere with a radius of 2.58 µm33, a rectangular box of 1 cm x 0.5 cm would need approximately 1.7 x 106 cells to form a monolayer. Care should be taken to ensure that there is no physical touching between the cells patched for mating and the two haploid cell grids. Since equal numbers of each type of haploid cells have to be mixed, 8.5 x 105 cells are added from each strain. The cell number is calculated based on OD measurements, considering 1 OD at 600 nm to be approximately equivalent to 1 x 107 cells34. For example, if the OD600 of the a haploid suspension is 0.17 and that of the α haploid is 0.11, the number of cells in 5 μL of each haploid suspension can be calculated. To ensure 8.5 x 105 cells of each haploid type, 1.25 μL of the a haploid and 1.93 μL of the α haploid suspensions are mixed. - Add the required volumes of both the haploids in a fresh and sterile 1.5 mL vial, and mix well using a pipette. The final volume of this suspension is generally around 6–8 µL. Patch this suspension in the center grid. Incubate the plate at 30 °C for 7 h, allowing the haploids sufficient time to mate (Figure 2E).
3. Plating of mixed cells on YPD agar
- After the incubation period of 7 h, scrape the cells from the center rectangle using a toothpick or a pipette tip, and dilute in 2 mL of sterile water. Then, spread the cell suspension on YPD agar to obtain single colonies. To determine the dilution factor necessary to obtain single colonies, measure the OD of the first tube into which the scraped cells are added. Specific dilution factors need to be determined for each cell type/strain being used.
NOTE: For example, an OD600 of 0.15 corresponds to 3 x 106 cells in a 2 mL suspension (considering 1 OD = 1 x 107 cells/mL). To obtain a few hundred colonies on the YPD plate, the cell suspension is serially diluted at 1:20 twice, and then 100 μL of the final dilution is used for spreading. - After plating, incubate the YPD plates at 30 °C for 36–48 h until there are single colonies. Ensure that a few hundred individual colonies are obtained from each mating experiment for screening so as to ensure statistical significance can be detected in the data (Figure 2F).
4. Screening for diploids using auxotrophic markers
- Determine what fraction of the colonies obtained are diploid. To identify the diploid colonies on the plate, transfer the single colonies by streaking them individually onto a double drop-out plate (2% glucose, 0.66% nitrogen base, 0.05% double drop-out amino acid mixture, and 2% agar) lacking the amino acids that the strains are auxotrophic for, as shown in Figure 2G. Incubate the plates at 30 °C for 48 h.
NOTE: The colonies can also be transferred onto the double drop-out plate using replica plating. In this study, tryptophan and leucine (trp− leu−) drop-out medium was used when quantifying the mating efficiency of the SK1AM strains, and tryptophan and uracil (trp− ura−) drop-out medium was used for the ScAM strains. Only the diploid colonies grow on the double drop-out plate as they have both the auxotrophic markers: TRP1 and LEU2 genes in the SK1AM strains and TRP1 and URA3 genes in the ScAM strains. - Additionally, streak or replica plate the colonies on single drop-out media (trp− or leu− or ura−) to quantify the frequency of each of the two kinds of haploid in the population.
- Calculate the mating efficiency, η, as follows:
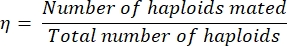 Eq (1)
Eq (1)
where the number of haploids mated is simply equal to twice the number of diploids identified on the double drop-out plate (since each diploid resulted from the mating of two haploids). The total number of haploids equals the sum of the number of haploids streaked plus twice the number of diploids streaked.
NOTE: For example, if only 60 colonies grow after streaking/replica plating 100 colonies on a double drop-out media, the mating efficiency can be quantified as 75% (as 60 x 2 haploids mated to form the 60 diploids, and 40 haploids did not mate).
Results
Quantification of the mating efficiency of the two mating types
The protocol described here was used to quantify the mating efficiency between two yeast strains-between SK1AMa and SK1AMα and between ScAMa and ScAMα (Figure 3A). In these experiments, the mating between the two haploids was repeated at least 12 times. In each of the repeats of the experiment, at least 100 colonies were streaked...
Discussion
The quantification of the mating efficiency in S. cerevisiae is essential for carrying out studies related to the genes involved in mating pathways or studying the influence of the external environment on mating behavior. In the past two decades, S. cerevisiae has also become a popular model to address questions related to speciation14,36,37,38. The presence of two mating type...
Disclosures
The authors declare that they have no competing interests in this work. The authors are happy to share the SK1-derived strains for all non-profit use.
Acknowledgements
This work was funded by a DBT/Wellcome Trust (India Alliance) grant (IA/S/19/2/504632) to S.S. P.N. is a Research Fellow supported by a DBT/Wellcome Trust (India Alliance) grant (IA/S/19/2/504632). A.M. is supported by the Council of Scientific and Industrial Research (CSIR), Government of India, as a Senior Research Fellow (09/087(0873)/2017-EMR-I). The authors thank Paike Jayadeva Bhat for discussions.
Materials
| Name | Company | Catalog Number | Comments |
| Adenine | Sigma Life Science | A8626 | |
| Agar Powder regular grade for bacteriology | SRL | 19661 (0140186) | |
| Ammonium Sulphate, Hi-AR | HiMedia | GRM1273 | |
| D-(+)-glucose | Sigma Life Science | G8270 | |
| Glass Petri plates | HiMedia | PW008 | 90 mm x 15 mm dimension |
| L-Arginine | Sigma Life Science | A8094 | |
| L-Aspartic acid | Sigma Life Science | A7219 | |
| L-Histidine monochloride monohydrate | Sigma Life Science | H5659 | |
| L-Isoleucine | Sigma Aldrich | I2752 | |
| L-Leucine | Sigma Life Science | L8912 | |
| L-Lysine | Aldrich | 62840 | |
| L-Methionine | Sigma Life Science | M5308 | |
| L-Phenylalanine | Sigma Life Science | P5482 | |
| L-Threonine | Sigma Aldrich | T8625 | |
| L-Tyrosine | Sigma Life Science | T8566 | |
| L-Valine | Sigma Life Science | V0513 | |
| Mating efficiency grid | 1 cm x 1.5 cm rectangular grid drawn on the Petri plate | ||
| Microcentrifuge tubes | Tarsons | 500010 | |
| Peptone | HiMedia | RM001 | |
| Uracil | Sigma Life Science | U0750 | |
| Yeast Extract Powder | HiMedia | RM027 | |
| Yeast Nitrogen Base w/o Amino acids and Ammonium Sulphate | BD Difco | 233520 |
References
- Neiman, A. M. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 189 (3), 737-765 (2011).
- Duina, A. A., Miller, M. E., Keeney, J. B. Budding yeast for budding geneticists: A primer on the Saccharomyces cerevisiae model system. Genetics. 197 (1), 33-48 (2014).
- Herskowitz, I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiological Reviews. 52 (4), 536-553 (1988).
- Erdman, S., Lin, L., Malczynski, M., Snyder, M. Pheromone-regulated genes required for yeast mating differentiation. Journal of Cell Biology. 140 (3), 461-483 (1998).
- Merlini, L., Dudin, O., Martin, S. G. Mate and fuse: How yeast cells do it. Open Biology. 3 (3), 130008 (2013).
- Gustin, M. C., Albertyn, J., Alexander, M., Davenport, K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 62 (4), 1264-1300 (1998).
- Bardwell, L. A walk-through of the yeast mating pheromone response pathway. Peptides. 26 (2), 339-350 (2005).
- Reid, B. J., Hartwell, L. H. Regulation of mating in the cell cycle of Saccharomyces cerevisiae. Journal of Cell Biology. 75, 355-365 (1977).
- Williams, T. C., Peng, B., Vickers, C. E., Nielsen, L. K. The Saccharomyces cerevisiae pheromone-response is a metabolically active stationary phase for bio-production. Metabolic Engineering Communications. 3, 142-152 (2016).
- Bagnat, M., Simons, K. Cell surface polarization during yeast mating. Proceedings of the National Academy of Sciences of the United States of America. 99 (22), 14183-14188 (2002).
- Trueheart, J., Boeke, J. D., Fink, G. R. Two genes required for cell fusion during yeast conjugation: Evidence for a pheromone-induced surface protein. Molecular and Cellular Biology. 7 (7), 2316-2328 (1987).
- Sniegowski, P. D., Dombrowski, P. G., Fingerman, E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Research. 1 (4), 299-306 (2002).
- Replansky, T., Koufopanou, V., Greig, D., Bell, G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends in Ecology and Evolution. 23 (9), 494-501 (2008).
- Greig, D. Reproductive isolation in Saccharomyces. Heredity. 102 (1), 39-44 (2009).
- Mayr, E. . Systematics and the Origin of Species, from the Viewpoint of a Zoologist. , (1999).
- Sprague, G. F. Assay of yeast mating reaction. Methods in Enzymology. 194, 77-93 (1991).
- McCaffrey, G., Clay, F. J., Kelsay, K., Sprague, G. F. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Molecular and Cellular Biology. 7 (8), 2680-2690 (1987).
- Valtz, N., Peter, M., Herskowitz, I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. Journal of Cell Biology. 131 (4), 863-873 (1995).
- Maclean, C. J., Greig, D. Prezygotic reproductive isolation between Saccharomyces cerevisiae and Saccharomyces paradoxus. BMC Evolutionary Biology. 8, 1 (2008).
- Murphy, H. A., Kuehne, H. A., Francis, C. A., Sniegowski, P. D. Mate choice assays and mating propensity differences in natural yeast populations. Biology Letters. 2 (4), 553-556 (2006).
- Leu, J. Y., Murray, A. W. Experimental evolution of mating discrimination in budding yeast. Current Biology. 16 (3), 280-286 (2006).
- Kim, J., Hirsch, J. P. A nucleolar protein that affects mating efficiency in Saccharomyces cerevisiae by altering the morphological response to pheromone. Genetics. 149 (2), 795-805 (1998).
- Jin, M., et al. Yeast dynamically modify their environment to achieve better mating efficiency. Science Signaling. 4 (186), (2011).
- Rogers, D. W., Denton, J. A., McConnell, E., Greig, D. Experimental evolution of species recognition. Current Biology. 25 (13), 1753-1758 (2015).
- McClure, A. W., Jacobs, K. C., Zyla, T. R., Lew, D. J. Mating in wild yeast: Delayed interest in sex after spore germination. Molecular Biology of the Cell. 29 (26), 3119-3127 (2018).
- Sena, E. P., Radin, D. N., Fogel, S. Synchronous mating in yeast. Proceedings of the National Academy of Sciences of the United States of America. 70 (5), 1373-1377 (1973).
- Werner-Washburne, M., Braun, E., Johnston, G. C., Singer, R. A. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiology Reviews. 57 (2), 383-401 (1993).
- Johnston, S. A., Hopper, J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proceedings of the National Academy of Sciences of the United States of America. 79 (22), 6971-6975 (1982).
- Mahilkar, A. . Study of metabolic specialization leading to speciation, using yeast as a model system. , (2021).
- Blank, T. E., Woods, M. P., Lebo, C. M., Xin, P., Hopper, J. E. Novel Gal3 proteins showing altered Gal80p binding cause constitutive transcription of Gal4p-activated genes in Saccharomyces cerevisiae. Molecular and Cellular Biology. 17 (5), 2566-2575 (1997).
- Rice, W. R., Hostert, E. E. Laboratory experiments on speciation: What have we learned in 40 years. Evolution. 47 (6), 1637-1653 (1993).
- White, N. J., Snook, R. R., Eyres, I. The past and future of experimental speciation. Trends in Ecology and Evolution. 35 (1), 10-21 (2020).
- Milo, R., Jorgensen, P., Moran, U., Weber, G., Springer, M. BioNumbers--The database of key numbers in molecular and cell biology. Nucleic Acids Research. 38, 750-753 (2010).
- Domitrovic, T., et al. Structural and functional study of YER067W, a new protein involved in yeast metabolism control and drug resistance. PLoS One. 5 (6), 11163 (2010).
- Lindegren, C. C., Spiegelman, S., Lindegren, G. Mendelian inheritance of adaptive enzymes. Proceedings of the National Academy of Sciences of the United States of America. 30 (11), 346-352 (1944).
- Dettman, J. R., Sirjusingh, C., Kohn, L. M., Anderson, J. B. Incipient speciation by divergent adaptation and antagonistic epistasis in yeast. Nature. 447 (7144), 585-588 (2007).
- Jhuang, H. Y., Lee, H. Y., Leu, J. Y. Mitochondrial-nuclear co-evolution leads to hybrid incompatibility through pentatricopeptide repeat proteins. EMBO Reports. 18 (1), 87-101 (2017).
- Lee, H. Y., et al. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 135 (6), 1065-1073 (2008).
- Pronk, J. T. Auxotrophic yeast strains in fundamental and applied research. Applied and Environmental Microbiology. 68 (5), 2095-2100 (2002).
- Madhani, H. D. . From a to α: Yeast as a Model for Cellular Differentiation. , (2007).
- Huxley, C., Green, E. D., Dunham, I. Rapid assessment of S. cerevisiae mating type by PCR. Trends in Genetics. 6 (8), 236 (1990).
- Rang, C., Galen, J. E., Kaper, J. B., Chao, L. Fitness cost of the green fluorescent protein in gastrointestinal bacteria. Canadian Journal of Microbiology. 49 (9), 531-537 (2003).
- Tusso, S., Nieuwenhuis, B. P. S., Weissensteiner, B., Immler, S., Wolf, J. B. W. Experimental evolution of adaptive divergence under varying degrees of gene flow. Nature Ecology and Evolution. 5 (3), 338-349 (2021).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved