Method Article
Fine Adjustment of Caenorhabditis elegans Orientation on Channeled Agar Pads for Imaging Neuroregeneration
In This Article
Summary
Here, we present a protocol for fabricating channeled agar pads using PDMS molds created from vinyl records. The channels enable users to finely orient Caenorhabditis elegans to improve imaging contrast and facilitate the comparison of structures. These capabilities are particularly useful in neuroregeneration studies.
Abstract
Caenorhabditis elegans is a model organism widely used for studying biological processes. Its transparency and small size make it ideal for imaging tissues, cells, and subcellular structures. Traditional flat agar pads for imaging C. elegans limit control over the animal's orientation, restricting views primarily to lateral perspectives. This limitation complicates the visualization of dorsal-ventral structures and reduces image clarity, especially in older animals with increased pigmentation and larger diameters. To overcome these challenges, we developed channeled agarose pads that allow precise control of animal orientation. These channels enable researchers to rotate and fix C. elegans in specified positions, facilitating the simultaneous imaging of multiple structures and improving image resolution by bringing target cells closer to the microscope objective. This is particularly useful for imaging regenerated neuronal fibers after surgery, which may grow in directions difficult to capture with traditional flat agar pads. This method is accessible, as fabricating channeled agar pads requires the same time and materials as flat pads, making it a practical option for most laboratories.
Introduction
The nematode Caenorhabditis elegans (C. elegans) is a widely utilized model organism for studying biological processes such as development and behavior due to its strongly conserved genetics with mammalian counterparts1. Researchers cultivate C. elegans on a nematode growth medium (NGM) plate in a tightly controlled environment that standardizes experimental conditions and allows for more precise attribution of results to experimental variables. The animal is transparent, enabling clear imaging of tissues, cells, and subcellular structures. C. elegans are small, with the adult animals being ~1 mm in length and 80 µm in diameter, which permits the placement of multiple animals on a single slide mount for imaging1. Their small size also allows a single image to capture the whole organism while resolving individual cells, which is crucial for visualizing the regeneration of neuron fibers. Synergistic to these advantages for microscopy, the rapid development, stereotyped anatomy, and facile genetics of C. elegans allow for large-scale studies1.
The typical procedure for imaging of cells in C. elegans was established decades ago and has largely remained unchanged. Researchers use flat agar pads on slides to mount and immobilize the animals for visualization of their cells2. A coverslip on top of the pad holds the animals in place and protects the lens of the microscope. Importantly, the coverslip has a high refractive index that increases the numerical aperture of captured light and improves imaging resolution. Additionally, the coverslip reduces distortion in light transmission. Thus, a coverslip improves the visualization of target cells and structures.
When used with a coverslip, however, flat agar pads often restrict animal orientation. C. elegans bend along their dorsal-ventral (DV) direction so they adopt a lateral orientation on a flat agar pad. However, even after immobilizing, straightening, and pre-rotating to a DV orientation, coverslip placement reverts nearly all animals into a lateral view (Figure 1). We believe this reorientation occurs because the coverslip pushes the protruding C. elegans vulva to the side. Younger animals without a vulva are more randomly oriented. This limited control over orientation is problematic for at least two reasons. First, it may not permit placing two structures in a single field of view (e.g., bilateral) which complicates their comparison. Second, setting the orientation is important for optimizing imaging quality, as structures of interest are generally best imaged when they are close to the objective. This is because objectives image a limited depth within the sample (i.e., working distance) and because the light from deeper locations experiences more scattering and absorption.
For C. elegans, the limited control over orientation is generally more serious with older animals. L1 to L4 age animals are smaller in diameter, making a higher fraction of the worm imageable. These larvae also have less pigmentation, resulting in reduced absorption of visible light and scattering of photons, which improves image clarity and resolution. Conversely, adult animals have a larger diameter and more pigmentation, which poses challenges for imaging in deeper z-planes since the coverslip affects the initial orientation.
In 2008, we introduced a strategy using agar pads with channels to overcome these challenges by maintaining the orientation of animals3,4. As described below, users rotate immobilized animals in the channels to a desired orientation by noting anatomical landmarks. The channels hold animals in depressions between elevated agar surfaces, minimizing force on the animal during coverslip placement and eliminating rotation of the animal. As the fabrication of channeled pads and common flat pads follow nearly the same procedure and require the same time, our technique is highly accessible. Many laboratories utilize our technique to immobilize C. elegans for studying anatomical features, observing development, and analyzing neuronal contributions to behavior5,6,7,8. The following section describes the entire process for making channeled agar, starting from a vinyl record which was cut into quarters using a hot knife. First, we describe the fabrication of the polydimethylsiloxane (PDMS) mold used to cast channeled agar. Then, we show how to fabricate the channeled agar step by step. The entire procedure is expected to take 3 h without expertise. Following the PDMS fabrication procedure, pads made with the mold will take the same time to make as typical agar pads (a few minutes).
Protocol
1. Preparing polydimethylsiloxane (PDMS) for fabrication
- Pour a 1:10 ratio of the fast cure agent to the base into a disposable weighing dish. Mix the uncured liquid for 45 s until the liquid is fully integrated and full of bubbles.
- Place the tray in a vacuum desiccator at a tilt. Cycle the pressure between -0.09 to -0.1 kPa 3x to allow air bubbles to surface.
- Keep the vacuum on for about an hour at a pressure range of -0.09 to -0.1 kPa until there are few remaining visible bubbles, which can be removed manually.
2. Fabricating PDMS mold of vinyl record
- Rinse the vinyl record and glass plate with deionized (DI) water. Ensure the absence of dust to obtain the best mold. Let the vinyl record air dry until used.
- Place a sheet of aluminum foil on top of the hot plate to catch excess PDMS.
- Put a glass slide on each end of the vinyl record to set the mold thickness to the width of the glass slides. This ensures uniform height when pressing the glass pane onto the uncured PDMS.
- Pour the uncured PDMS liquid on one side of the vinyl record.
- Observe bubbles using light reflection off liquid surface and remove them with a pipette tip. Tilt the glass plate and bring the plate down slowly allowing air to escape, preventing bubbles from being introduced.
- Cure the PDMS at 80 °C for 35 min.
- Remove the vinyl record from the hot plate and let it cool. Carefully peel the PDMS from the vinyl record to avoid tearing.
- Use a new glass slide as a guide to cut the PDMS with a sharp razor to create a channeled agar mold.
3. Fabricating channeled agar pads with NaN3
NOTE: The procedure and materials for flat agar pads were previously established9.This procedure is very similar.
- Add 0.6 g of agar and 30 mL of NGM liquid9 stock into a flask. Add a stir bar and heat on a hot plate at 120 °C.
- Add 120 µL of NaN3 stock (1 M stock concentration) after the gel is melted.
- Wash the PDMS mold with water and let it air dry. Place the PDMS mold between two sets of slide pairs.
- Pipette agar up and down to warm the pipette tip. Pipette 300 µL of melted agar onto the PDMS mold, manually spreading agar across channels, as slide placement will spread agar along channels.
- Place a microscope slide directly onto the agar, resting on the slides on the sides. Remove the slide after the agar cools.
4. Mounting C. elegans
- Once the slides cool, lift the slide straight off the mold without shearing.
- Move animals into the channels after they are immobilized by NaN3
- Rotate the animals to the desired orientation.
- Place a coverslip on the pad. Tilt the pad and push down lightly from one side to the other to push air to the side and avoid creating bubbles.
Results
Fabricating PDMS mold
Cycling the vacuum pressure during mold fabrication punctures and removes air bubbles from the uncured PDMS, ensuring a bubble-free mold. As shown in Figure 3D shows, bubbles rise to the surface and puncture. Bubble-free molds can produce clean channels.
Solidification of PDMS mold
When placing the glass pane on the vinyl record, avoid creating air gaps. These gaps can lead to uneven thickness or surface defects. Bubbles, if present, can be detected visually. Pressing the glass pane from one side to the other displaces air, minimizing bubble formation and ensuring an even mold thickness shown in Figure 4A.
Using channeled agar pads for fine orientation of C. elegans
The channeled agar pads allow precise orientation of C. elegans. We set the animal orientation by lightly rolling the animal with a platinum wire pick, while verifying orientation using landmarks, such as the vulva or the S-shaped intestine (Figure 5A,C). We confirm the orientation under a fluorescent dissecting microscope before transferring animals to an inverted microscope shown in Figure 5B,D.
Assessing regeneration of neuron fibers with channeled agar pads
Channeled agar pads improve the imaging quality of neuronal regeneration by optimally orienting the animal. Positioning regenerated neuron fibers closer to the objective lens reduces light scattering and absorption, enhancing image clarity. Figure 5E,F show that placing the regeneration site closer to the objective enhances imaging quality, facilitating more accurate analysis of neuronal regrowth.
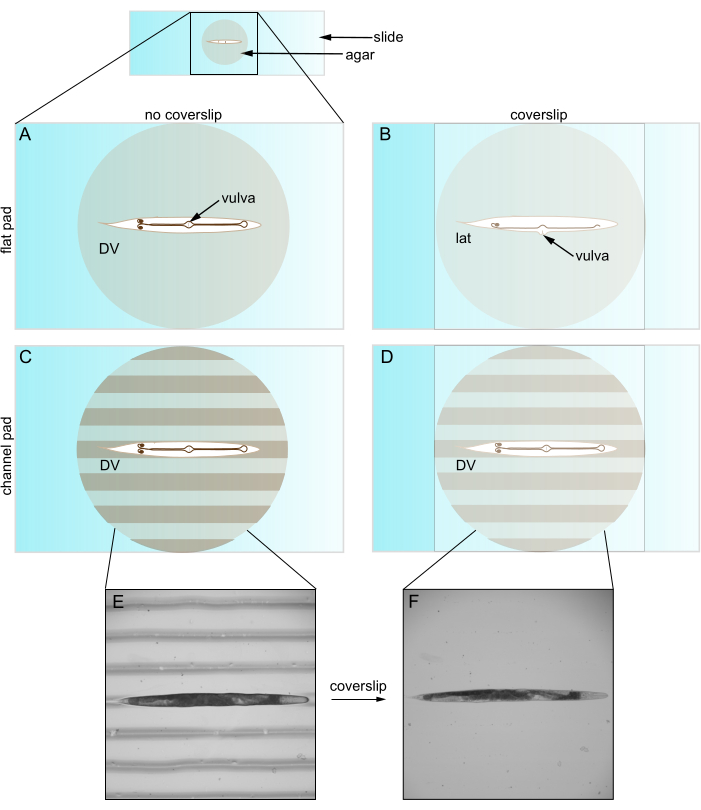
Figure 1: Caenorhabditis elegans on flat and channeled pads. (A) Animal on flat agar pad in DV orientation; vulva faces coverslip. (B) Coverslip introduction pushes vulva to side, forcing animal into lateral orientation. (C) Animal on channeled agar pad in DV orientation. (D) Animal remains in DV orientation after coverslip introduction. (E) Brightfield image of roller mutant on channeled agar pad in DV orientation. (F) Brightfield image with coverslip, same animal as E remains in DV orientation. Abbreviations: DV = dorsal-ventral; lat = lateral. Please click here to view a larger version of this figure.
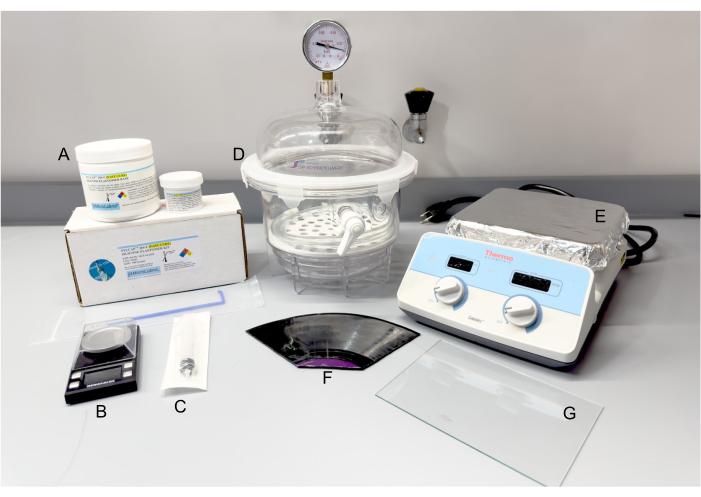
Figure 2: Materials and equipment for PDMS fabrication. (A) SYLCAP 284-F Silicone Elastomer Kit. (B) Scale. (C) Plastic syringe. (D) Vacuum desiccator. (E) Hot plate. (F) 12-inch long-playing vinyl record, break apart as needed. (G) Glass pane. Abbreviation: PDMS = polydimethylsiloxane. Please click here to view a larger version of this figure.
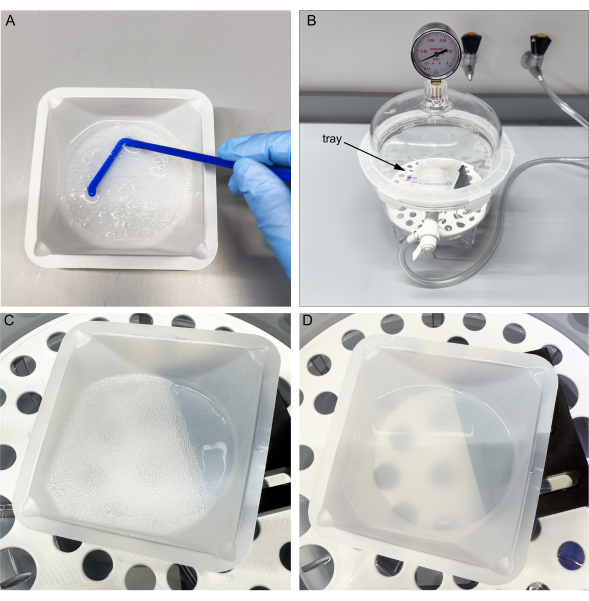
Figure 3: Mixing and vacuuming uncured PDMS. (A) Mix the base and curing agent in the tray. (B) Place tray in a vacuum chamber. (C) Cycle vacuum three times to allow bubbles to surface. (D) After one hour, most bubbles popped. Note improved transmission through PDMS. Please click here to view a larger version of this figure.
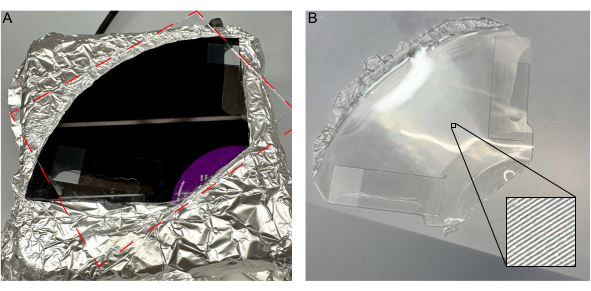
Figure 4: Introduction of a glass pane to uncured PDMS without bubbles. (A) Introduction of glass pane on vinyl record without bubbles. (B) Solidified PDMS mold removed from the record, with inset detailing channels. Please click here to view a larger version of this figure.
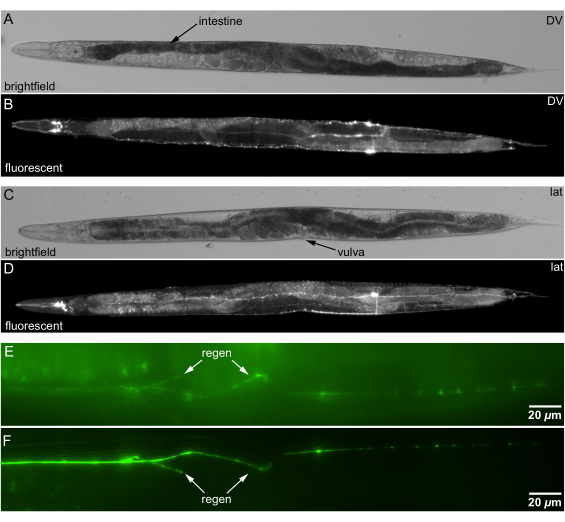
Figure 5: Brightfield and fluorescence images of C. elegans mounted on channeled agar. (A) Brightfield image of an animal in bilateral DV orientation. Anatomical landmark for DV orientation: spiral S-shaped intestine (darker tissue indicated by arrow) crossing midline ~60% of the distance from nose to tail. (B) Fluorescence image of neurons in DV orientation, same animal as A. (C) Brightfield image of an animal in lat orientation. Anatomical landmark for lat orientation: vulva facing side. (D) Fluorescence image of neurons in lat orientation, same animal as C. (E) Orientation with regeneration (arrow) in deeper (far) side of an animal. (F) Orientation with regeneration in shallower side animal closer to objective; regeneration clearer (arrow). Scale bars = 20 µm. Abbreviations: DV = dorsal-ventral; lat = lateral. Please click here to view a larger version of this figure.
Discussion
The primary advantage of channeled agar pads is their ability to maintain the orientation of animals for imaging. The channels fit adult animals, allowing controlled rotation that is maintained upon coverslip introduction. We have successfully imaged wild-type, roller, and dumpy animals. We properly orient animals using anatomical landmarks, which allows imaging and comparison of multiple structures in one field of view and brings target cells closer to the surface. In particular, this rotation facilitates visualization in the dorsal or ventral (bilateral views), which allow comparison of the left and right side of the animal.
Previously, the visualization and comparison of these bilateral structures would require 3D z-stacks and potential normalization of signals to account for reduced transmission at greater depths. Thus, the defined orientation enabled by channeled agar allows for more consistent imaging across multiple worms and should allow improved measurements. This is particularly helpful for postsurgery assessment of regenerated neuronal fibers, which can grow in directions that are difficult to image, such as the circumferential2.
Additionally, the channeled agar reduces stress on animals because they are placed in a depression which alleviates coverslip pressure and promotes normal physiology5. While flaccid strains may twist during mounting, the channels encourage a linear animal configuration and rotation of the entire animal. In a different implementation, the patterning can also constrain animal movement to reduce the amount of anesthetic required to image or observe the animal8.
Finally, the channeled agar technique is also highly accessible. After mold fabrication, creating channeled agar pads requires roughly the same amount of time, expertise, and cost as making regular flat agar pads. All components needed to create the PDMS mold are common to many academic labs or bought online. The durability of PDMS molds make them a cost-effective option for labs, capable of withstanding extensive use over time. PDMS molds can last for years and hundreds of uses. We continue to use a 17-year-old mold with moderate degradation. We recommend storing the molds at room temperature in a container to keep them free from debris, which can be washed away with deionized water if necessary.
Despite these advantages, there are several limitations associated with our technique. The force of the pick used to place animals on the pad can deform the channels, potentially affecting the positioning of the animal. As with any fine manipulation, positioning and rotating the animal to the desired orientation can be time-consuming, which increases the animal's exposure to anesthetic and potentially induces stress. Channels can also trap air and form bubbles during the placement of the coverslip, which can interfere with the clarity of imaging by scattering light. We discuss ways to minimize bubbles in the protocol. Our experience indicates that most of the disadvantages of channeled agar are largely avoided by a modicum of training and practice.
Future directions for optimizing these channels include developing designs tailored to specific experimental needs, including different sizes and shapes for different ages and phenotypes of C. elegans. Fabricating customized channel sizes by nanofabrication10 may reduce physiological impact on animals compared to generic channels5. In conclusion, the implementation of channeled agar pads enables high-resolution imaging of obscure structures in older animals. These pads maintain the orientation of C. elegans, leading to enhanced imaging quality and detailed visualization, while reducing stress on the animals due to the supportive structure of the channels.
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
Funding was provided by NIH R56NS128413. We acknowledge Noah Joseph (Northeastern Bioengineering Department) for assistance with processing vinyl records. We thank members of Chung Laboratory for insightful comments on the manuscript.
Materials
| Name | Company | Catalog Number | Comments |
| 125 mL Erlenmeyer flask | Corning | 4980-125 | |
| 33 1/2 RPM or 12-inch long playing (LP) vinyl record | N/A | N/A | Individual channel depth: 50 µm, width: 200 µm |
| 30 mL plastic syringe | Fisher Scientific | 50-793-817 | |
| 90% Platinum, 10% Iridium Wire | Tritech Research | PT-9010 | used to create platinum pick |
| aluminum foil | Amazon | N/A | |
| Calcium chloride | Fisher Scientific | AAL131910B | |
| glass pane | N/A | N/A | 7 in x 5 in glass pane taken from frame |
| glass pipette | Fisher Scientific | 13-678-20A | used to create platinum pick |
| hot plate | Thermofisher | SP88857104 | |
| Magnesium Sulfate | Fisher Scientific | AA3333736 | |
| microscope slides | Mckesson | 938360 | |
| molecular biology grade agar | Benchmark Scientific | A1705 | |
| Potassium Phosphate dibasic | Fisher Scientific | P290-500 | |
| Potassium Phosphate monobasic | Fisher Scientific | P285-500 | |
| Sodium Azide | Fisher Scientific | AAJ2161022 | |
| Sodium Chloride | Fisher Scientific | BP358-212 | |
| square cover glass | Fisher Scientific | 12-541-016 | |
| SYLCAP 284-F (Fast Cure) Silicone Elastomer Encapsulant Kit, Transparent, Optically Clear, 10:1 Mix, 500 Gm/ML (0.5 Kg), Faster Than Sylgard 184 and Similar | MicroLubrol | N/A | |
| vacuum desiccator | Fisher Scientific | 08-648-100 | |
| weighing dish | Fisher Scientific | 01-549-750 |
References
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics. 77 (1), 71-94 (1974).
- Bargmann, C. I., Avery, L. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 48, 225-250 (1995).
- Zhang, M., et al. A self-regulating feed-forward circuit controlling C-elegans egg-laying behavior. Curr Biol. 18 (19), 1445-1455 (2008).
- Chung, S. H., Mazur, E. Femtosecond laser ablation of neurons in C. elegans for behavioral studies. Appl Phys A-Mater Sci Process. 96 (2), 335-341 (2009).
- Zellag, R. M., et al. CentTracker: a trainable, machine-learning-based tool for large-scale analyses of Caenorhabditis elegans germline stem cell mitosis. Mol Biol Cell. 32 (9), 915-930 (2021).
- Huang, Y. C., et al. A single neuron in C. elegans orchestrates multiple motor outputs through parallel modes of transmission. Curr Biol. 33 (20), 4430-4445.e6 (2023).
- Rasmussen, N. R., Dickinson, D. J., Reiner, D. J. Ras-dependent cell fate decisions are reinforced by the RAP-1 small GTPase in Caenorhabditis elegans. Genetics. 210 (4), 1339-1354 (2018).
- Rivera Gomez, K., Schvarzstein, M. Immobilization of nematodes for live imaging using an agarose pad produced with a vinyl record. MicroPubl Biol. 2018, (2018).
- Chung, S. H., Clark, D. A., Gabel, C. V., Mazur, E., Samuel, A. D. T. The role of the AFD neuron in C. elegans thermotaxis analyzed using femtosecond laser ablation. BMC Neuroscience. 7 (1), 30(2006).
- Gerhold Abigail, R., et al. Investigating the regulation of stem and progenitor cell mitotic progression by in situ imaging. Curr Biol. 25 (9), 1123-1134 (2015).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved