需要订阅 JoVE 才能查看此. 登录或开始免费试用。
Method Article
RNA-seq数据中替代剪接和聚腺苷酸化的鉴定
摘要
选择性剪接(AS)和替代聚腺苷酸化(APA)扩大了转录本亚型及其产物的多样性。在这里,我们描述了生物信息学协议,以分析批量RNA-seq和3'末端测序测定,以检测和可视化不同实验条件下变化的AS和APA。
摘要
除了对RNA-Seq进行典型分析以测量实验/生物学条件下的差异基因表达(DGE)外,RNA-seq数据还可用于探索外显子水平的其他复杂调控机制。选择性剪接和聚腺苷酸化通过产生不同的亚型来调节转录后水平的基因表达,在基因的功能多样性中起着至关重要的作用,并且将分析限制在整个基因水平上可能会错过这一重要的调控层。在这里,我们演示了详细的分步分析,以使用Bioconductor和其他封装和功能(包括DEXSeq,Limma封装的diffSplice和rMATS)来识别和可视化不同条件下的差异外显子和聚腺苷酸化位点的使用。
引言
多年来,RNA-seq已被广泛用于估计差异基因表达和基因发现1。此外,它还可用于估计由于表达不同亚型的基因而导致的不同外显子水平使用情况,从而有助于更好地了解转录后水平的基因调控。大多数真核基因通过交替剪接(AS)产生不同的亚型,以增加mRNA表达的多样性。AS事件可分为不同的模式:跳过完全外显子(SE),其中("盒式")外显子与其侧翼内含子一起从转录本中完全去除;当外显子两端存在两个或多个剪接位点时,备选(供体)5'剪接位点选择(A5SS)和备选方案3'(受体)剪接位点选择(A3SS);当内含子保留在成熟的mRNA转录本中时保留内含子(RI)和相互排除外显子使用(MXE),其中一次只能保留两个可用外显子中的一个2,3。替代聚腺苷酸化(APA)在使用替代聚(A)位点从单个转录本产生多种mRNA亚型4的基因表达中也起着重要作用。大多数聚腺苷酸化位点(pA)位于3'非翻译区域(3'UTR),产生具有不同3'UTR长度的mRNA亚型。由于 3' UTR 是识别调控元件的中心枢纽,因此不同的 3' UTR 长度会影响 mRNA 的定位、稳定性和翻译5。有一类 3' 末端测序测定经过优化,可检测 APA,这些APA在协议6的细节上有所不同。此处描述的管道是为 PolyA-seq 设计的,但可以适用于所述的其他协议。
在这项研究中,我们提出了一系列差异外显子分析方法7,8 (图1),可分为两大类:基于外显子(DEXSeq9,diffSplice10)和基于事件的(复制转录本剪接的多变量分析(rMATS)11)。基于外显子的方法将单个外显子条件下的倍数变化与整体基因折叠变化的度量进行比较,以调用差异表达的外显子使用情况,并由此计算AS活性的基因水平测量。基于事件的方法使用外显子-内含子跨越结读取来检测和分类特定的剪接事件,例如外显子跳跃或内含子保留,并在输出3中区分这些AS类型。因此,这些方法为AS12,13的完整分析提供了补充观点。我们选择了DEXSeq(基于DESeq214 DGE封装)和diffSplice(基于Limma10 DGE封装)进行研究,因为它们是差分拼接分析中使用最广泛的软件包之一。rMATS被选为基于事件的分析的常用方法。另一种流行的基于事件的方法是MISO(亚型混合物)1。对于APA,我们采用基于外显子的方法。
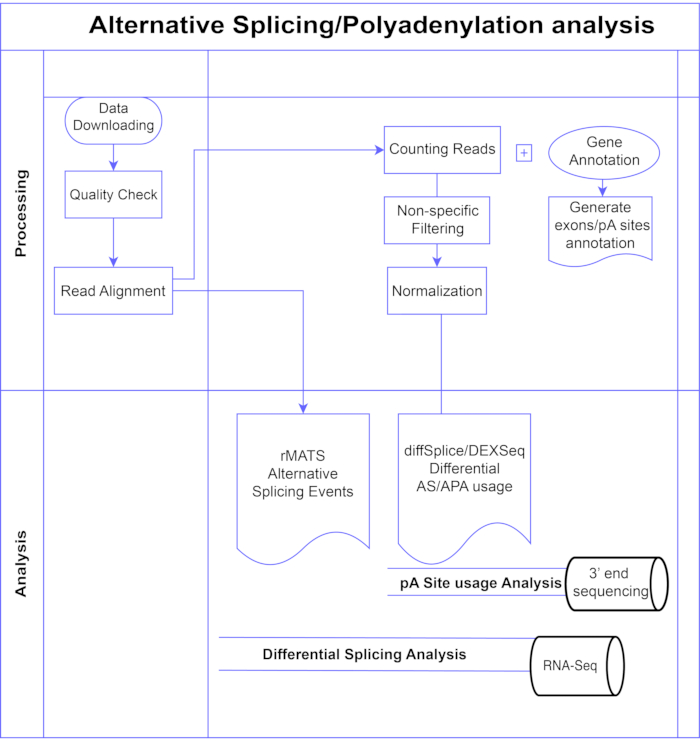
图1.分析管道。 分析中使用的步骤的流程图。步骤包括:获取数据,执行质量检查和读取对齐,然后使用已知外显子,内含子和pA位点的注释对读取进行计数,过滤以去除低计数和标准化。使用diffSplice/DEXSeq方法分析PolyA-seq数据以寻找替代pA位点,使用diffSplice/DEXseq方法分析外显子水平的替代剪接的体RNA-Seq,并使用rMATS分析AS事件。 请点击此处查看此图的大图。
本调查中使用的RNA-seq数据是从基因表达综合(GEO)(GSE138691)15获得的。我们使用本研究的小鼠RNA-seq数据,分为两个条件组:野生型(WT)和肌盲样1型敲除(Mbnl1 KO),每个重复三个。为了证明差异聚腺苷酸化位点的使用分析,我们获得了小鼠胚胎成纤维细胞(MEFs)PolyA-seq数据(GEO加入GSE60487)16。数据有四个条件组:野生型(WT),肌肉盲样1型/2型双敲除(Mbnl1 / 2 DKO),Mbnl 1 / 2 DKO与Mbnl3敲低(KD)和Mbnl1 / 2 DKO与Mbnl3对照(Ctrl)。每个条件组由两个仿行组成。
| 加入全球环境展望 | SRA 运行编号 | 示例名称 | 条件 | 复制 | 组织 | 测 序 | 读取长度 | |
| 核糖核酸序列 | GSM4116218 | SRR10261601 | Mbnl1KO_Thymus_1 | Mbnl1 淘汰赛 | 代表 1 | 胸腺 | 配对端 | 100 基点 |
| GSM4116219 | SRR10261602 | Mbnl1KO_Thymus_2 | Mbnl1 淘汰赛 | 代表 2 | 胸腺 | 配对端 | 100 基点 | |
| GSM4116220 | SRR10261603 | Mbnl1KO_Thymus_3 | Mbnl1 淘汰赛 | 代表 3 | 胸腺 | 配对端 | 100 基点 | |
| GSM4116221 | SRR10261604 | WT_Thymus_1 | 野生型 | 代表 1 | 胸腺 | 配对端 | 100 基点 | |
| GSM4116222 | SRR10261605 | WT_Thymus_2 | 野生型 | 代表 2 | 胸腺 | 配对端 | 100 基点 | |
| GSM4116223 | SRR10261606 | WT_Thymus_3 | 野生型 | 代表 3 | 胸腺 | 配对端 | 100 基点 | |
| 3P-序列 | GSM1480973 | SRR1553129 | WT_1 | 野生型(WT) | 代表 1 | 小鼠胚胎成纤维细胞 (MEF) | 单端 | 40 基点 |
| GSM1480974 | SRR1553130 | WT_2 | 野生型(WT) | 代表 2 | 小鼠胚胎成纤维细胞 (MEF) | 单端 | 40 基点 | |
| GSM1480975 | SRR1553131 | DKO_1 | Mbnl 1/2 双淘汰赛 (DKO) | 代表 1 | 小鼠胚胎成纤维细胞 (MEF) | 单端 | 40 基点 | |
| GSM1480976 | SRR1553132 | DKO_2 | Mbnl 1/2 双淘汰赛 (DKO) | 代表 2 | 小鼠胚胎成纤维细胞 (MEF) | 单端 | 40 基点 | |
| GSM1480977 | SRR1553133 | DKOsiRNA_1 | Mbnl 1/2 双敲除与 Mbnl 3 siRNA (KD) | 代表 1 | 小鼠胚胎成纤维细胞 (MEF) | 单端 | 40 基点 | |
| GSM1480978 | SRR1553134 | DKOsiRNA_2 | Mbnl 1/2 双敲除与 Mbnl 3 siRNA (KD) | 代表 2 | 小鼠胚胎成纤维细胞 (MEF) | 单端 | 36 基点 | |
| GSM1480979 | SRR1553135 | DKONTsiRNA_1 | Mbnl 1/2 双敲除与非靶向 siRNA (Ctrl) | 代表 1 | 小鼠胚胎成纤维细胞 (MEF) | 单端 | 40 基点 | |
| GSM1480980 | SRR1553136 | DKONTsiRNA_2 | Mbnl 1/2 双敲除与非靶向 siRNA (Ctrl) | 代表 2 | 小鼠胚胎成纤维细胞 (MEF) | 单端 | 40 基点 |
表 1.用于分析的RNA-Seq和PolyA-seq数据集摘要。
研究方案
1. 安装分析中使用的工具和 R 包
- Conda 是一个流行且灵活的包管理器,允许在所有平台上方便地安装包及其依赖项。使用"Anaconda"(conda包管理器)安装"conda",可用于安装分析所需的工具/包。
- 根据 https://www.anaconda.com/products/individual#Downloads 的系统要求下载"Anaconda",并按照图形安装程序中的提示进行安装。通过在 Linux 命令行中键入以下内容,使用 'conda' 安装所有必需的软件包。
conda install -c daler sratoolkit
conda install -c conda-forge parallel
conda install -c bioconda star bowtie fastqc rmats rmats2sashimiplot samtools fasterq-dump cutadapt bedtools deeptools - 若要下载协议中使用的所有 R 包,请在 R 控制台(通过键入"R"在 Linux 命令行上启动)或 Rstudio 控制台中键入以下代码。
bioc_packages<- c("DEXSeq", "Rsubread", "EnhancedVolcano", "edgeR", "limma", "maser","GenomicRanges")
packages<- c("magrittr", "rtracklayer", "tidyverse", "openxlsx", "BiocManager")
#Install if not already installed
installed_packages<-packages%in% rownames(installed.packages())
installed_bioc_packages<-bioc_packages%in% rownames(installed.packages())
if(any(installed_packages==FALSE)) {
install.packages(packages[!installed_packages],dependencies=TRUE)
BiocManager::install(packages[!installed_bioc_packages], dependencies=TRUE)
}
注意:在此计算协议中,命令将作为 R 笔记本文件(扩展名为".Rmd"),R 代码文件(扩展名为".R"),或 Linux Bash shell 脚本(扩展名为".sh"的文件)。R 笔记本 (Rmd) 文件应使用 File| 在 RStudio 中打开打开 File...,然后通过单击右上角的绿色箭头以交互方式运行单个代码块(可能是 R 命令或 Bash shell 命令)。R代码文件可以通过在RStudio中打开来运行,或者在Linux命令行上通过前缀"Rscript"来运行,例如Rscript example.R.Shell脚本通过在脚本前面加上"sh"命令在Linux命令行上运行,例如.sh example.sh。
2. 使用 RNA-seq 进行选择性剪接 (AS) 分析
- 数据下载和预处理
注意:下面注释的代码片段可在补充代码文件中找到"AS_analysis_RNASeq.Rmd",以交互方式遵循各个步骤,并且还作为 bash 脚本提供,以便在 Linux 命令行上批量运行(什downloading_data_preprocessing.sh).- 下载原始数据。
- 使用 SRA 工具包 (v2.10.8) 中的"预取"命令从序列读取存档 (SRA) 下载原始数据17。在以下命令中按顺序提供 SRA 入藏 ID,以使用 GNU 并行实用程序18 并行下载它们。要将入藏 ID 的 SRA 文件从 SRR10261601 并行下载到 SRR10261606,请在 Linux 命令行上使用以下方法。
seq 10261601 10261606 | parallel prefetch SRR{} - 使用SRA工具包中的"fastq-dump"功能从存档中提取fastq文件。使用 GNU 并行并一起给出所有 SRA 文件的名称。
parallel -j 3 fastq-dump --gzip --skip-technical --read-filter pass --dumpbase --split-e --clip --origfmt {} ::: - 在 Linux 命令行使用以下方法从 www.ensembl.org 下载小鼠(基因组组装 GRCm39)的参考基因组和注释。
wget -nv -O annotation.gtf.gz http://ftp.ensembl.org/pub/release-103/gtf/mus_musculus/Mus_musculus.GRCm39.103.gtf.gz \ && gunzip -f annotation.gtf.gz
wget -nv -O genome.fa.gz http://ftp.ensembl.org/pub/release-103/fasta/mus_musculus/dna/Mus_musculus.GRCm39.dna.primary_assembly.fa.gz \ && gunzip -f genome.fa.gz
GTF=$(readlink -f annotation.gtf)
GENOME=$(readlink -f genome.fa)
- 使用 SRA 工具包 (v2.10.8) 中的"预取"命令从序列读取存档 (SRA) 下载原始数据17。在以下命令中按顺序提供 SRA 入藏 ID,以使用 GNU 并行实用程序18 并行下载它们。要将入藏 ID 的 SRA 文件从 SRR10261601 并行下载到 SRR10261606,请在 Linux 命令行上使用以下方法。
- 预处理和映射读取到基因组组装
- 质量管理。使用 FASTQC 评估原始读取的质量(FASTQ 质量检查 v0.11.9)19。创建一个输出文件夹,并在多个输入 fasta 文件上并行运行 fastqc。此步骤将为每个样品生成质量报告。在进行进一步分析之前,请检查报告以确保读取质量是可接受的。(请参阅用户手册以了解 https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ 中的报告)
mkdir fastqc_out
parallel "fastqc {} -o fastqc_out" ::: $RAW_DATA/*.fastq.gz
注意:如有必要,使用"cutadapt"20 或"trimmomatic"21 执行适配器修整,以去除侧翼接头的测序,这因RNA片段大小和读取长度而异。在此分析中,我们跳过了此步骤,因为受影响的读取比例很小。 - 读取对齐方式。预处理的下一步包括将读段映射到参考基因组。首先,使用 STAR22 的"genomeGenerate"功能构建参考基因组的索引,然后将原始读数与参考对齐(或者,可从 STAR 网站获得预构建索引,并可直接用于对齐)。在 Linux 命令行运行以下命令。
#Build STAR index
GDIR=STAR_indices
mkdir $GDIR
STAR --runMode genomeGenerate --genomeFastaFiles $GENOME --sjdbGTFfile $GTF --runThreadN 8 --genomeDir $GDIR
ODIR=results/mapping
mkdir -p $ODIR
#Align reads to the genome
for fq1 in $RAW_DATA/*R1.fastq.gz;
do
fq2=$(echo $fq1| sed 's/1.fastq.gz/2.fastq.gz/g');
OUTPUT=$(basename ${fq1}| sed 's/R1.fastq.gz//g');
STAR --genomeDir $GDIR \
--runThreadN 12 \
--readFilesCommand zcat \
--readFilesIn ${fq1}${fq2}\
--outFileNamePrefix $ODIR\/${OUTPUT} \
--outSAMtype BAM SortedByCoordinate \
--outSAMunmapped Within \
--outSAMattributes Standard
Done
注意:读取对齐后,STAR 对准器将为每个样本生成和排序 BAM(二进制对齐图)文件。在继续执行后续步骤之前,必须对 Bam 文件进行排序。
- 质量管理。使用 FASTQC 评估原始读取的质量(FASTQ 质量检查 v0.11.9)19。创建一个输出文件夹,并在多个输入 fasta 文件上并行运行 fastqc。此步骤将为每个样品生成质量报告。在进行进一步分析之前,请检查报告以确保读取质量是可接受的。(请参阅用户手册以了解 https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ 中的报告)
- 下载原始数据。
- 准备外显子注释。
- 运行补充代码文件"prepare_mm_exon_annotation.R",下载的GTF(基因转移格式)格式的注释以准备注释。若要运行,请在 Linux 命令行中键入以下内容。
Rscript prepare_mm_exon_annotation.R annotation.gtf
注意:GTF 文件包含不同亚型的多个外显子条目。此文件用于"折叠"每个外显子的多个转录本 ID。这是定义外显子计数箱的重要步骤。
- 运行补充代码文件"prepare_mm_exon_annotation.R",下载的GTF(基因转移格式)格式的注释以准备注释。若要运行,请在 Linux 命令行中键入以下内容。
- 计数读取。下一步是计算映射到不同转录本/外显子的读取次数。请参阅补充文件:"AS_analysis_RNASeq.Rmd"。
- 加载所需的库:
packages<- c("Rsubread","tidyverse", "magrittr", "EnhancedVolcano", "edgeR","openxlsx")
invisible(lapply(packages, library, character.only=TRUE)) - 加载从上一步 (2.2) 获得的已处理注释文件。
load("mm_exon_anno.RData") - 读取在步骤 2.2.2 中获得的所有 bam 文件作为"featureCounts"的输入以计算读取次数。通过首先列出以 .bam 结尾的目录中的每个文件来读取包含 bam 文件的文件夹。使用 Rsubread 包中的"featureCounts",该包将 bam 文件和处理后的 GTF 注释(参考)作为输入,以生成与每个特征关联的计数矩阵,其中行表示外显子(特征)和列表示样本。
countData <- dir("bams", pattern=".bam$", full.names=T) %>%
featureCounts(annot.ext=anno,
isGTFAnnotationFile=FALSE,
minMQS=0,useMetaFeatures=FALSE,
allowMultiOverlap=TRUE,
largestOverlap=TRUE,
countMultiMappingReads=FALSE,
primaryOnly=TRUE,
isPairedEnd=TRUE,
nthreads=12) - 接下来,执行非特异性过滤以去除低表达的外显子("非特异性"表示过滤中不使用实验条件信息,以避免选择偏差)。使用"edgeR"包23 中的 cpm 函数将数据从原始规模转换为百万分之一 (cpm),并在至少三个样本中保持计数大于可设置阈值的外显子(对于此数据集使用 1 cpm)。同时去除只有一个外显子的基因。
# Non-specific filtering: Remove the exons with low counts
isexpr<- rownames(countData$counts)[rowSums(cpm(countData$counts)>1) >=3]
countData$counts<-countData$counts[rownames(countData$counts) %in%isexpr, ]
anno<-anno%>% filter(GeneID%in% rownames(countData$counts))
# Remove genes with only 1 site and NA in geneIDs
dn<-anno%>%group_by(GeneID)%>%summarise(nsites=n())%>% filter(nsites>1&!is.na(GeneID))
anno<-anno%>% filter(GeneID%in%dn$GeneID)
countData$counts<-countData$counts[rownames(countData$counts) %in%anno$GeneID, ]
注意:使用不同数据时,请检查 featureCount 的必需参数,例如,对于单端读取,请设置"isPairedEnd = FALSE"。请参阅 RSubread 用户指南以选择数据选项,并参阅下面的"讨论"部分。
- 加载所需的库:
- 差分拼接和外显子使用分析。我们描述了此步骤的两种替代方案:DEXSeq 和 DiffSplice。两者都可以使用并给出类似的结果。为了保持一致性,如果您更喜欢用于 DGE 的 DESeq2 软件包,请选择 DEXSeq,并使用 DiffSplice 进行基于 Limma 的 DGE 分析。请参阅补充文件:"AS_analysis_RNASeq.Rmd".
- 使用 DEXSeq 软件包进行差分外显子分析。
- 加载库并创建示例表以定义实验设计。
library(DEXSeq)
sampleTable<-data.frame(row.names= c("Mbnl1KO_Thymus_1", "Mbnl1KO_Thymus_2", "Mbnl1KO_Thymus_3", "WT_Thymus_1", "WT_Thymus_2", "WT_Thymus_3"), condition= rep(c("Mbnl1_KO", "WT"),c(3,3)), libType= rep(c("paired-end")))
注意:行名应与 featureCounts 用于计算读取计数的 bam 文件名一致。样本表包含每个样本的详细信息,其中包括:库类型和条件。这是定义用于检测差异使用情况的对比度或测试组所必需的。 - 准备外显子信息文件。在下一步中创建 DEXSeq 对象时,需要 GRanges(基因组范围)对象 (https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html) 形式的外显子信息作为输入。将基因 ID 与读取计数匹配以创建外显子信息对象。
exoninfo<-anno[anno$GeneID%in% rownames(countData$counts),]
exoninfo<-GRanges(seqnames=anno$Chr,
ranges=IRanges(start=anno$Start, end=anno$End, width=anno$Width),strand=Rle(anno$Strand))
mcols(exoninfo)$TranscriptIDs<-anno$TranscriptIDs
mcols(exoninfo)$Ticker<-anno$Ticker
mcols(exoninfo)$ExonID<-anno$ExonID
mcols(exoninfo)$n<-anno$n
mcols(exoninfo)$GeneID<-anno$GeneID
transcripts_l= strsplit(exoninfo$TranscriptIDs, "\\,")
save(countData, sampleTable, exoninfo, transcripts_l, file="AS_countdata.RData") - 使用 DEXSeqDataSet 函数创建 DEXSeq 对象。DEXSeq 对象将读取计数、外显子特征信息和样本信息收集在一起。使用步骤 3 中生成的读取计数和从上一步获得的外显子信息从计数矩阵创建 DEXSeq 对象。sampleData 参数采用定义样本(及其属性:库类型和条件)的数据帧输入,"design"使用 sampleData 使用模型公式表示法生成用于差异测试的设计矩阵。请注意,一个重要的相互作用项,条件:外显子,表明落在特定外显子上的基因的读取分数取决于实验条件,即存在AS。有关为更复杂的实验设计设置模型公式的完整说明,请参阅 DEXSeq 文档。对于特征信息,需要外显子ID,相应的基因和转录本。
dxd<-DEXSeqDataSet(countData$counts,sampleData=sampleTable, design=~sample+exon+condition:exon,featureID=exoninfo$ExonID,groupID=exoninfo$GeneID,featureRanges=exoninfo, transcripts=transcripts_l) - 归一化和离散估计。接下来,使用以下命令在样品之间执行归一化并估计数据的方差,这是由于 RNA-seq 离散性质和生物变异性的泊松计数噪声。
dxd %<>% estimateSizeFactors %>% estimateDispersions %T>% plotDispEsts - 测试差异使用情况。估计变异后,测试每个基因的差异外显子使用情况并生成结果。
dxd%<>%testForDEU%>%estimateExonFoldChanges(fitExpToVar=
"condition")#Estimate fold changes
dxr=DEXSeqResults(dxd) - 使用以下命令可视化选定基因的剪接事件。
plotDEXSeq(dxr,"Wnk1", displayTranscripts=TRUE, splicing=TRUE,legend
=TRUE,cex.axis=1.2,cex=1.3,lwd=2)
检查 R 笔记本文件"AS_analysis_RNASeq.Rmd"以生成感兴趣的基因的其他图,并生成不同阈值的火山图。
- 加载库并创建示例表以定义实验设计。
- 使用来自 Limma 的差分拼接来识别差分拼接。按照 R 笔记本文件"AS_analysis_RNASeq.Rmd".在继续之前,请确保已按照步骤 2.1-2.3 准备输入文件。
- 加载库
library(limma)
library(edgeR) - 非特定筛选。提取在 2.3 中获得的读取计数矩阵。使用edgeR包中的"DGEList"函数创建特征列表,其中行代表基因,列代表样本。
mycounts=countData$counts
#Change the rownames of the countdata to exon Ids instead of genes for unique rownames.
rownames(mycounts) = exoninfo$ExonID
dge<-DGEList(counts=mycounts)
#Filtering
isexpr<- rowSums(cpm(dge)>1) >=3
dge<-dge[isexpr,,keep.lib.sizes=FALSE]
#Extract the exon annotations for only transcripts meeting non-specific filter
exoninfo=anno%>% filter(ExonID%in% rownames(dge$counts))
#Convert the exoninfo into GRanges object
exoninfo1<-GRanges(seqnames=exoninfo$Chr,
ranges=IRanges(start=exoninfo$Start, end=exoninfo$End, width=exoninfo$Width),strand=Rle(exoninfo$Strand))
mcols(exoninfo1)$TranscriptIDs<-exoninfo$TranscriptIDs
mcols(exoninfo1)$Ticker<-exoninfo$Ticker
mcols(exoninfo1)$ExonID<-exoninfo$ExonID
mcols(exoninfo1)$n<-exoninfo$n
mcols(exoninfo1)$GeneID<-exoninfo$GeneID
transcripts_l= strsplit(exoninfo1$TranscriptIDs, "\\,")
注意:作为非特异性过滤步骤,计数按 cpm 过滤 < n 个样本中的 x 个中的 1 个,其中 x 是任何条件下的最小重复次数。此示例数据的 n = 6 和 x = 3。 - 使用"edgeR"包中的"calcNormFactors"函数使用修剪的 M 值平均值(TMM 归一化方法)24 对样本的计数进行归一化 它将计算比例因子以调整库大小。
dge<-calcNormFactors(dge) - 使用步骤 2.4.1.1 中生成的 sampleTable 并创建设计矩阵。设计矩阵表征了设计。有关更高级实验设计的设计矩阵的详细信息,请参阅 Limma 用户指南 (https://www.bioconductor.org/packages/devel/bioc/vignettes/limma/inst/doc/usersguide.pdf) 第 8 章和第 9 章。
Treat<- factor(sampleTable$condition)
design<- model.matrix(~0+Treat)
colnames(design) <- levels(Treat) - 拟合每个外显子的线性模型。运行"limma"包的"voom"功能来处理RNA-seq数据以估计方差并生成精度权重以校正泊松计数噪声,并将外显子水平计数转换为每百万对数2计数(logCPM)。然后使用"lmfit"函数运行线性建模,将线性模型拟合到每个外显子的表达数据中。使用"eBayes"函数计算拟合模型的经验贝叶斯统计量,以检测微分外显子表达。接下来,为感兴趣的实验比较定义对比矩阵。使用"contrasts.fit"获取每对比较的系数和标准误差。
v<-voom(dge,design,plot=FALSE)
fit<-lmFit(v,design)
fit<-eBayes(fit)
colnames(fit)
cont.matrix<-makeContrasts(
Mbnl1_KO_WT=Mbnl1_KO-WT,
levels=design)
fit2<-contrasts.fit(fit,cont.matrix) - 差分拼接分析。在拟合模型上运行"diffSplice"以测试野生型和敲除型之间基因外显子使用的差异,并使用"topSplice"函数探索排名靠前的结果:test="t"给出AS外显子的排名,test="simes"给出基因的排名。
ex<-diffSplice(fit2,geneid=exoninfo$GeneID,exonid=exoninfo$ExonID)
ts<-topSplice(ex,n=Inf,FDR=0.1, test="t", sort.by="logFC")
tg<-topSplice(ex,n=Inf,FDR=0.1, test="simes") - 可视化。使用"plotSplice"函数绘制结果,在基因参数中给出感兴趣的基因。保存按日志排序的顶部结果 将变化折叠成一个对象并生成火山图以展示外显子。
plotSplice(ex,geneid="Wnk1", FDR=0.1)
#Volcano plot
EnhancedVolcano(ts,lab=ts$ExonID,selectLab= head((ts$ExonID),2000), xlab= bquote(~Log[2]~'fold change'), x='logFC', y='P.Value', title='Volcano Plot', subtitle='Mbnl1_KO vs WT (Limma_diffSplice)', FCcutoff=2, labSize=4,legendPosition="right", caption= bquote(~Log[2]~"Fold change cutoff, 2; FDR 10%"))
- 加载库
- 使用 rMATS
- 确保在工作目录中使用 conda 或 github (https://github.com/Xinglab/rmats-turbo/releases/download/v4.1.1/rmats_turbo_v4_1_1.tar.gz) 安装了最新版本的 rMATS v4.1.1(也称为 rMATS turbo,因为处理时间和内存要求更少)。遵循"AS_analysis_RNASeq.Rmd"中的第 4.3 节。
- 转到包含映射后获得的 bam 文件的文件夹,并根据 rMAT 的要求,通过复制以","分隔的 bam 文件的名称(以及路径)来准备文本文件。应在 Linux 命令行运行以下命令:
mkdir rMATS_analysis
cd bams/
ls -pd "$PWD"/*| grep "WT"| tr '\n'','> Wt.txt
ls -pd "$PWD"/*| grep "Mb"| tr '\n'','> KO.txt
mv *.txt ../rMATS_analysis - 使用上一步中生成的两个输入文件以及 2.1.1.3 中获取的 GTF 文件运行 rmats.py。这将生成一个输出文件夹"rmats_out",其中包含分别描述每个拼接事件的统计数据(p 值和包含水平)的文本文件。
python rmats-turbo/rmats.py --b1 KO.txt --b2 Wt.txt --gtf annotation.gtf -t paired --readLength 50 --nthread 8 --od rmats_out/ --tmp rmats_tmp --task pos
注意:还需要 GTF 文件形式的参考注释。检查参数是否为单端数据,并相应地更改 -t 选项。 - 探索 rMATS 结果。使用Bioconductor包'maser'25 来探索rMATS结果。将结点和外显子计数 (JCEC) 文本文件加载到"激射器"对象中,并通过包括每个拼接事件至少五个平均读取来根据覆盖范围过滤结果。
library(maser)
mbnl1<-maser("/rmats_out/", c("WT","Mbnl1_KO"), ftype="JCEC")
#Filtering out events by coverage
mbnl1_filt<-filterByCoverage(mbnl1,avg_reads=5) - 可视化 rMATS 结果。使用"maser"包中的"topEvents"函数,以 10% 的错误发现率 (FDR) 和拼接百分比 (deltaPSI) 的最小 10% 变化选择显著的拼接事件。接下来,检查感兴趣的单个基因(样本基因-Wnk1)的基因事件,并绘制该基因的每个剪接事件的PSI值。通过指定事件类型生成火山图。
#Top splicing events at 10% FDR
mbnl1_top<-topEvents(mbnl1_filt,fdr=0.1, deltaPSI=0.1)
mbnl1_top
#Check the gene events for a particular gene
mbnl1_wnk1<-geneEvents(mbnl1_filt,geneS="Wnk1", fdr=0.1, deltaPSI=0.1)
maser::display(mbnl1_wnk1,"SE")
plotGenePSI(mbnl1_wnk1,type="SE", show_replicates
=TRUE)
volcano(mbnl1_filt,fdr=0.1, deltaPSI=0.1,type="SE")
+xlab("deltaPSI")+ylab("Log10 Adj. Pvalue")+ggtitle("Volcano Plot of exon skipping events") - 使用"rmats2shahimiplot"包以文本文件的形式为使用rMATS获得的拼接事件结果生成生鱼片图。在 Linux 命令行运行 python 脚本。
python ./src/rmats2sashimiplot/rmats2sashimiplot.py --b1 ../bams/WT_Thymus_1.bam,../bams/WT_Thymus_2.bam,../bams/WT_Thymus_3.bam --b2 ../bams/Mbnl1KO_Thymus_1.bam,../bams/Mbnl1KO_Thymus_2.bam,../bams/Mbnl1KO_Thymus_3.bam -t SE -e ../rMATS_analysis/rmats_out/SE.MATS.JC.txt --l1 WT --l2 Mbnl1_KO --exon_s 1 --intron_s 5 -o ../rMATS_analysis/rmats2shasmi_output
注意:此过程可能非常耗时,因为它将为事件文件中的所有结果生成生鱼片图。从"maser"中选择topEvents函数显示的顶级结果(基因名称和外显子),并可视化相应的生鱼片图。
- 使用 DEXSeq 软件包进行差分外显子分析。
3. 使用 3' 末端测序的替代聚腺苷酸化 (APA) 分析
- 数据下载和预处理
注意: 请参阅补充 R 笔记本文件"APA_analysis_3PSeq_notebook。rmd"表示用于数据下载和预处理步骤的完整命令,或在 Linux 命令行上运行补充 bash 文件 "APA_data_downloading_preprocessing.sh"。- 使用入藏 ID(1553129 至 1553136)从 SRA 下载数据。
- 调整适配器和反向补码以获得检测链序列。
注意:此步骤特定于所使用的PolyA-seq测定。 - 使用领结对齐器26将读取映射到小鼠基因组组装。
- 准备 pA 站点注释。
注意:pA网站注释文件的处理首先使用补充R笔记本文件"APA_analysis_3PSeq_notebook。Rmd"(2.1 - 2.6),然后使用 bash 文件 "APA_annotation_preparation.sh"。- 从 PolyASite 2.0 数据库下载 pA 站点注释6.
- 选择 pA 位点注释以保留 3'-非翻译区域 (UTR) pA 位点,这些位点注释为末端外显子 (TE) 或注释末端外显子 (DS) 下游 1000 nt 以进行下游分析。
- 获得pA位点峰。锚定在每个pA切割位点,并使用bedtools和deeptools27,28可视化平均读取覆盖率。结果表明,映射读段的峰主要分散在切割位点上游~60 bp内(图5和补充图5)。因此,pA位点的坐标从注释文件扩展到其裂解位点上游60 bp。根据所使用的特定3'末端测序方案,此步骤需要针对PolyA-seq以外的检测进行优化。
- 计数读取
- 准备 pA 站点注释文件。
anno<- read.table(file= "flanking60added.pA_annotation.bed",
stringsAsFactors=FALSE, check.names=FALSE, header=FALSE, sep="")
colnames(anno) <- c("chrom", "chromStart", "chromEnd", "name", "score", "strand", "rep", "annotation", "gene_name", "gene_id")
anno<- dplyr::select(anno,name,chrom, chromStart,chromEnd, strand,gene_id,gene_name,rep)
colnames(anno) <- c("GeneID", "Chr", "Start", "End", "Strand", "Ensembl", "Symbol", "repID") - 应用"特征计数"以获取原始计数。将计数表另存为 RData 文件"APA_countData.Rdata",以便使用不同的工具进行 APA 分析。
countData<- dir("bamfiles", pattern="sorted.bam$", full.names=TRUE) %>%
# Read all bam files as input for featureCounts
featureCounts(annot.ext=anno, isGTFAnnotationFile= FALSE,minMQS=0,useMetaFeatures= TRUE,allowMultiOverlap=TRUE, largestOverlap= TRUE,strandSpecific=1, countMultiMappingReads =TRUE,primaryOnly= TRUE,isPairedEnd= FALSE,nthreads=12)%T>%
save(file="APA_countData.Rdata")
注意:请注意更改"功能计数"函数中列出的任何参数。修改"strandSpecific"参数,以确保它与所使用的3'末端测序测定的测序方向一致(根据经验,在基因组浏览器中可视化正负链上的基因数据将阐明这一点)。 - 对计数数据应用非特定筛选。滤波可以显著提高差分pA站点使用测试中的统计鲁棒性。首先,我们去除了那些只有一个pA位点的基因,无法定义差异化的pA位点使用。其次,我们应用基于覆盖率的非特异性过滤:计数通过 cpm 过滤,在 n 个样本中小于 x 中的 1 个,其中 x 是任何条件下的最小重复次数。对于此示例数据,N = 8 和 x = 2。
load(file= "APA_countData.Rdata")# Skip this step if already loaded
# Non-specific filtering: Remove the pA sites not differentially expressed in the samples
countData<-countData$counts%>%as.data.frame%>% .[rowSums(edgeR::cpm(.)>1) >=2, ]
anno%<>% .[.$GeneID%in% rownames(countData), ]
# Remove genes with only 1 site and NA in geneIDs
dnsites<-anno%>%group_by(Symbol)%>%summarise(nsites=n())%>% filter(nsites>1&!is.na(Symbol))
anno<-anno%>% filter(Symbol%in%dnsites$Symbol)
countData<-countData[rownames(countData) %in%anno$GeneID, ]
- 准备 pA 站点注释文件。
- 使用 DEXSeq 和 diffSplice 管道进行差异聚腺苷酸化位点使用分析。
- 使用 DEXSeq 软件包
注意:由于无法为 DEXSeq 管道定义对比矩阵,因此必须分别执行每个两个实验条件的差异 APA 分析。以条件WT和条件DKO的差异APA分析为例进行,以解释该过程。请参阅补充文件"APA_analysis_3PSeq_notebook。马币",用于本节的分步工作流程和其他对比度的差异APA分析。- 加载库并创建示例表以定义实验设计。
c("DEXSeq", "GenomicRanges") %>% lapply(library, character.only=TRUE) %>%invisible
sampleTable1<- data.frame(row.names= c("WT_1","WT_2","DKO_1","DKO_2"),
condition= c(rep("WT", 2), rep("DKO", 2)),
libType= rep("single-end", 4)) - 使用Bioconductor包GRanges准备pA位点信息文件。
# Prepare the GRanges object for DEXSeqDataSet object construction
PASinfo <- GRanges(seqnames = anno$Chr,
ranges = IRanges(start = anno$Start, end = anno$End),strand = Rle(anno$Strand))
mcols(PASinfo)$PASID<-anno$repID
mcols(PASinfo)$GeneEns<-anno$Ensembl
mcols(PASinfo)$GeneID<-anno$Symbol
# Prepare the new feature IDs, replace the strand information with letters to match the current pA site clusterID
new.featureID <- anno$Strand %>% as.character %>% replace(. %in% "+", "F") %>% replace(. %in% "-", "R") %>% paste0(as.character(anno$repID), .) - 使用步骤 3.3 中生成的读取计数和从上一步获得的 pA 站点信息来创建 DEXSeq 对象。
# Select the read counts of the condition WT and DKO
countData1<- dplyr::select(countData, SRR1553129.sorted.bam, SRR1553130.sorted.bam, SRR1553131.sorted.bam, SRR1553132.sorted.bam)
# Rename the columns of countData using sample names in sampleTable
colnames(countData1) <- rownames(sampleTable1)
dxd1<-DEXSeqDataSet(countData=countData1,
sampleData=sampleTable1,
design=~sample+exon+condition:exon,
featureID=new.featureID,
groupID=anno$Symbol,
featureRanges=PASinfo) - 通过定义 DEXSeq 对象中的条件级别来定义对比度对。
dxd1$condition<- factor(dxd1$condition, levels= c("WT", "DKO"))
# The contrast pair will be "DKO - WT" - 归一化和离散估计。与RNA-seq数据类似,对于3'末端测序数据,使用"估计大小因子"函数在样品之间执行归一化(每个样品的比率的列中位数),并使用"估计分散"函数估计数据的变化,然后使用"plotDispEsts"函数可视化色散估计结果。
dxd1 %<>% estimateSizeFactors %>% estimateDispersions %T>% plotDispEsts - 使用函数"testForDEU"对每个基因进行差异pA位点使用测试,然后使用函数"estimateExonFoldChanges"估计pA位点使用率的倍数变化。使用"DEXSeqResults"函数检查结果,并将"FDR < 10%"设置为显著差异 pA 位点的标准。
dxd1 %<>% testForDEU %>% estimateExonFoldChanges(fitExpToVar = "condition")
dxr1 <- DEXSeqResults(dxd1)
dxr1
mcols(dxr1)$description
table(dxr1$padj<0.1) # Check the number of differential pA sites (FDR < 0.1)
table(tapply(dxr1$padj<0.1, dxr1$groupID, any)) # Check the number of gene overlapped with differential pA site - 使用函数"plotDEXSeq"生成的差异APA图和函数"增强火山"生成的火山图可视化差异pA站点使用结果。
# Select the top 100 significant differential pA sites ranked by FDR
topdiff.PAS<- dxr1%>%as.data.frame%>%rownames_to_column%>%arrange(padj)%$%groupID[1:100]
# Apply plotDEXSeq for the visualization of differential polyA usage
plotDEXSeq(dxr1,"S100a7a", legend=TRUE, expression=FALSE,splicing=TRUE, cex.axis=1.2, cex=1.3,lwd=2)
# Apply perGeneQValue to check the top genes with differential polyA site usage
dxr1%<>% .[!is.na(.$padj), ]
dgene<- data.frame(perGeneQValue= perGeneQValue (dxr1)) %>%rownames_to_column("groupID")
dePAS_sig1<-dxr1%>% data.frame() %>%
dplyr::select(-matches("dispersion|stat|countData|genomicData"))%>%
inner_join(dgene)%>%arrange(perGeneQValue)%>%distinct()%>%
filter(padj<0.1)
# Apply EnhancedVolcano package to visualise differential polyA site usage
"EnhancedVolcano"%>% lapply(library, character.only=TRUE) %>%invisible
EnhancedVolcano(dePAS_sig1, lab=dePAS_sig1$groupID, x='log2fold_DKO_WT',
y='pvalue',title='Volcano Plot',subtitle='DKO vs WT',
FCcutoff=1,labSize=4, legendPosition="right",
caption= bquote(~Log[2]~"Fold change cutoff, 1; FDR 10%"))
- 加载库并创建示例表以定义实验设计。
- 使用差异拼接包。请参阅补充 R 笔记本文件"APA_analysis_3PSeq_notebook。Rmd",用于本节的分步工作流。
- 定义差异 pA 使用情况分析的目标对比度。
注意: 此步骤应在构造和处理 DGEList 对象之后执行,该对象包含在 R 笔记本文件"APA_analysis_3PSeq_notebook。啪��contrast.matrix<-makeContrasts(DKO_vs_WT=DKO-
WT,Ctrl_vs_DKO=Ctrl-DKO,
KD_vs_Ctrl=KD-Ctrl,KD_vs_DKO=KD-DKO,levels=design)
fit2<-fit%>%contrasts.fit(contrast.matrix)%>%eBayes
summary(decideTests(fit2))
ex<-diffSplice(fit2,geneid=anno$Symbol,exonid=new.featureID)
topSplice(ex) #Check the top significant results with topSplice - 使用函数"plotSplice"的差分APA图和函数"增强火山"的火山图可视化感兴趣的对比结果(此处为"DKO - WT")。请参阅 R 笔记本文件"APA_analysis_3PSeq_notebook.Rmd" 4.2.7 - 4.2.9 用于其他对比对的可视化。
sig1<-topSplice(ex,n=Inf,FDR=0.1,coef=1, test="t", sort.by="logFC")
sig1.genes<-topSplice(ex,n=Inf,FDR=0.1,coef=1, test="simes")
plotSplice(ex, coef=1,geneid="S100a7a", FDR = 0.1)
plotSplice(ex,coef=1,geneid="Tpm1", FDR = 0.1)
plotSplice(ex,coef=1,geneid="Smc6", FDR = 0.1)
EnhancedVolcano(sig1, lab=sig1$GeneID,xlab= bquote(~Log[2]~'fold change'),
x='logFC', y='P.Value', title='Volcano Plot', subtitle='DKO vs WT',
FCcutoff=1, labSize=6, legendPosition="right")
- 定义差异 pA 使用情况分析的目标对比度。
- 使用 DEXSeq 软件包
结果
运行上述分步工作流程后,AS和APA分析输出和代表性结果以表格和数据图的形式生成,生成如下。
如:
AS分析的主要输出(差异拼接的补充表1;DEXSeq的表2)是显示不同条件差异用法的外显子列表,以及显示其一个或多个组成外显子的显着整体剪接活性的基因列表,按统计学显着性排名。补充表1,选项卡2显示了显着的外显子?...
讨论
在这项研究中,我们评估了基于外显子和基于事件的方法,以检测批量RNA-Seq和3'末端测序数据中的AS和APA。基于外显子的AS方法既产生差异表达的外显子列表,又产生按总体基因水平差异剪接活性的统计显着性排序的基因水平排名(表1-2,4-5)。对于diffSplice包,差异用法是通过在外显子水平上拟合加权线性模型来确定的,以估计外显子的差异对数倍数变化与同一基因内其他外显子的平...
披露声明
作者没有什么可透露的。
致谢
这项研究得到了澳大利亚研究委员会(ARC)未来奖学金(FT16010043)和澳大利亚国立大学期货计划的支持。
材料
| Name | Company | Catalog Number | Comments |
| Not relevent for computational study |
参考文献
- Katz, Y., Wang, E. T., Airoldi, E. M., Burge, C. B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature Methods. 7 (12), 1009-1015 (2010).
- Wang, Y., et al. Mechanism of alternative splicing and its regulation. Biomedical Reports. 3 (2), 152-158 (2015).
- Mehmood, A., et al. Systematic evaluation of differential splicing tools for RNA-seq studies. Briefings in Bioinformatics. 21 (6), 2052-2065 (2020).
- Movassat, M., et al. Coupling between alternative polyadenylation and alternative splicing is limited to terminal introns. RNA Biology. 13 (7), 646-655 (2016).
- Tian, B., Manley, J. L. Alternative polyadenylation of mRNA precursors. Nature Reviews Molecular Cell Biology. 18 (1), 18-30 (2017).
- Herrmann, C. J., et al. PolyASite 2.0: a consolidated atlas of polyadenylation sites from 3' end sequencing. Nucleic Acids Research. 48 (1), 174-179 (2020).
- Liu, R., Loraine, A. E., Dickerson, J. A. Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems. BMC Bioinformatics. 15 (1), 364 (2014).
- Conesa, A., et al. A survey of best practices for RNA-seq data analysis. Genome Biology. 17 (1), 13 (2016).
- Anders, S., Reyes, A., Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Research. 22 (10), 2008-2017 (2012).
- Ritchie, M. E., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 43 (7), 47 (2014).
- Shen, S., et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proceedings of the National Academy of Sciences. 111 (51), 5593-5601 (2014).
- Mehmood, A., et al. Systematic evaluation of differential splicing tools for RNA-seq studies. Briefings in bioinformatics. 21 (6), 2052-2065 (2020).
- Kanitz, A., et al. Comparative assessment of methods for the computational inference of transcript isoform abundance from RNA-seq data. Genome biology. 16 (1), 1-26 (2015).
- Love, M. I., Huber, W., Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 15 (12), 550 (2014).
- Sznajder, L. J., et al. Loss of MBNL1 induces RNA misprocessing in the thymus and peripheral blood. Nature Communications. 11, 1-11 (2020).
- Batra, R., et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Molecular Cell. 56 (2), 311-322 (2014).
- Leinonen, R., Sugawara, H., Shumway, M., et al. The sequence read archive. Nucleic acids research. 39, 19-21 (2010).
- Tange, O. . GNU parallel-the command-line power tool. 36, 42-47 (2011).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 17 (1), 10-12 (2011).
- Bolger, A. M., Lohse, M., Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30 (15), 2114-2120 (2014).
- Dobin, A., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29 (1), 15-21 (2013).
- Robinson, M. D., McCarthy, D. J., Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26 (1), 139-140 (2010).
- Robinson, M. D., Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 11 (3), 25 (2010).
- Veiga, D. F. T. maser: Mapping Alternative Splicing Events to pRoteins. R package version 1.4.0. , (2019).
- Langmead, B., Trapnell, C., Pop, M., Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 10 (13), 25 (2009).
- Quinlan, A. R., Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26 (6), 841-842 (2010).
- Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A., Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic acids research. 42 (1), 187-191 (2014).
- Merino, G. A., Conesa, A., Fernández, E. A. A benchmarking of workflows for detecting differential splicing and differential expression at isoform level in human RNA-seq studies. Briefings in bioinformatics. 20 (2), 471-481 (2019).
- Chhangawala, S., Rudy, G., Mason, C. E., Rosenfeld, J. A. The impact of read length on quantification of differentially expressed genes and splice junction detection. Genome biology. 16 (1), 1-10 (2015).
- Conesa, A., et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016).
- Trapnell, C., et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7 (3), 562-578 (2012).
- Li, B., Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12, 323 (2011).
- Bray, N. L., Pimentel, H., Melsted, P., Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 34 (5), 525-527 (2016).
转载和许可
请求许可使用此 JoVE 文章的文本或图形
请求许可探索更多文章
This article has been published
Video Coming Soon
版权所属 © 2025 MyJoVE 公司版权所有,本公司不涉及任何医疗业务和医疗服务。