Method Article
The Importance of Correct Protein Concentration for Kinetics and Affinity Determination in Structure-function Analysis
W tym Artykule
Podsumowanie
We apply label-free protein interaction analysis using Biacore X100 for structure-function analysis of the binding of several cystatin B mutants to papain through kinetic characterization. Calibration-free concentration analysis (CFCA) measures the concentration of protein with retained binding activity without the need for a standard curve. We show that confirmation of concentrations using CFCA increases the reliability of the kinetic analysis and that kinetic constants can reliably be determined even if the activity of a recombinant protein is reduced.
Streszczenie
Protokół

Figure 1. The three-dimensional structure of the cystatin B complex (blue) with papain (yellow). Mutated cystatin B residues are shown in red.
1. Principles of Label-free Interaction Analysis using Biacore Systems
In a typical label-free binding experiment using a Biacore system, a biomolecule termed the 'ligand' is attached to the surface of a sensor chip. A system of flow-channels brings its binding partner, termed the 'analyte', into contact with the chip surface, where detection takes place. When the analyte binds to the ligand, the resulting change in accumulating mass at the surface is detected by surface plasmon resonance (SPR). The SPR response is proportional to the amount of binding analyte.
Since the binding is measured in real time, the kinetic association and dissociation rate constants for a specific interaction can be determined. From these constants, it is possible to calculate the affinity as the equilibrium dissociation constant. It is also possible to calculate affinity from steady state binding data. A similar methodology can also be used to determine the concentration of a protein that specifically binds to a ligand on the surface.
2. Analyzing kinetic properties of a protein-protein interaction
In Biacore X100, kinetic analysis can be performed using single-cycle kinetics. In a single-cycle kinetics experiment, a concentration series of the analyte is injected in a single analysis cycle without regeneration of the surface in between injections. Therefore, single-cycle kinetics enables kinetic analysis when it is difficult to find suitable regeneration conditions.
Once the data for a kinetic experiment has been collected, Biacore X100 evaluation software generates the values of ka, kd, and KD by fitting the data to an interaction model.
3. Approaches to Determining Protein Concentration
Analyzing the interactions between biomolecules is important for understanding their function. Characterizing the binding of proteins to other proteins, to nucleic acid, or to small molecules is fundamental to biochemical research, and finds use in many other areas including drug discovery.
For accurate measurement of the interaction kinetics between two interacting proteins it is essential to know the concentration of specifically binding protein in the experimental sample that is used as analyte. A spectrophotometer reading of A280 or colorimetric assays such as the one employing Bradford's reagent is commonly used to determine total protein concentration. However, protein impurities can affect the result. More importantly, both active and inactive forms of the protein are included in the total protein concentration. Particularly in the case of recombinant proteins, which may be inactive due to incorrect folding, it is important to determine the percentage of specifically binding protein in the sample.
In Biacore X100, concentration related to specific binding activity can either be determined by comparison of binding response levels to a calibration curve derived from a known standard, or by using the more recently introduced methodology Calibration-Free Concentration Analysis (CFCA). CFCA does not rely on to a standard. Therefore, CFCA is particularly useful in the case of studying mutant forms of proteins, where standards are usually not available.
In a CFCA experiment, the initial binding rate is measured at different flow rates under conditions when diffusion of sample to the chip surface is rate-limiting. The diffusion coefficient of the analyte, the dimensions of the flow cell and the flow rates are taken into account when calculating the specific binding concentration from the initial binding rate1,2.
In a kinetics experiment, the concentration of the analyte is used in the calculation of the kinetic association rate constant and the affinity from the experimental data. CFCA and kinetic measurement in Biacore systems both rely on the same interaction properties. Therefore, using the specific binding concentration determined by Biacore analysis, rather than total protein concentration, increases the reliability of the results.
4. Using Biacore analysis to Characterize the Interaction Between Cystatin B and Papain
Mammalian cystatin B is a reversible, competitive and tight-binding protein inhibitor of papain-like cysteine proteinases, primarily cathepsin B, H, K, L and S. These proteins are mainly involved in nonselective intracellular protein degradation. Cystatins are presumed to protect cells and tissues from inappropriate proteolysis by these enzymes. They also inactivate cysteine proteinases from parasites and viruses and may participate in the defense against invasion of such infectious agents. In addition, cystatins inhibit several plant cysteine proteinases, such as papain, which is frequently used as a model enzyme in structure-function studies. The three-dimensional structure of the cystatin B complex with papain3 shows that the interaction between the two proteins is dominated by hydrophobic contacts, which, from the inhibitor side, are provided by the N- and C-terminal ends and two hairpin loops (Figure 1). In this study, we examine the importance of the C-terminal end and the second binding loop of cystatin B for binding to papain by using bovine cystatin B variants containing point mutations in areas of interaction with papain. We first determine the specific binding concentration of the four cystatin B variants used, as well as that of the wild type protein. Once the specific binding concentrations of active cystatin B are determined, the rate and affinity constants of wild type and mutant variants of cystatin B binding to papain are measured using Biacore X100.
5. Instrument and Reagents
- The cystatin B variants Cys3Ser/His75Gly, Cys3Ser/Leu73Gly, Cys3Ser/Tyr97Ala and Cys3Ser are produced as previously described4. All mutants contain an additional substitution of cysteine to serine at position 3, to prevent the formation of disulfide-linked inactive dimers of cystatin B.
- The papain target is also prepared as previously described5. It is S-(methylthio) papain (MMTS-papain) having a methylthio group attached to Cys25 in the active cleft, rendering the protease catalytically inactive.
- Biacore X100 with Biacore X100 Plus Package is used to measure and analyze the specific binding concentrations and binding kinetics.
- MMTS-papain is immobilized on Sensor Chip CM5 using Biacore Amine Coupling Kit.
- The runs are performed at 25°C, and the buffer is 0.01 M Hepes at pH 7.4, 0.15 M NaCl, 0.0034 M EDTA, and 0.05% polysorbate 20.
- Between each variant of cystatin B, the sensor surface is regenerated with 20 mM NaOH for 30 seconds at a flow rate of 10 μl/min.
6.Immobilization of the Ligand MMTS-papain
Covalent attachment of MMTS-papain for CFCA analysis
CFCA relies on measurement of binding rate under conditions where the rate is limited by diffusion of analyte molecules to the surface (mass transport limitation). This is favored by high immobilization levels of the ligand. Immobilization of MMTS-papain was set up and run using the immobilization wizard of Biacore X100 Control software.
- Activate the surface in flow cell 2 by injection of a mixture of succinimide (NHS) and carbodiimide (EDC) for 7 minutes at a flow rate of 10 μl/min. Flow cell 1 is left unmodified in order to be used as a reference surface.
- Inject MMTS-papain at 50μg/ml in sodium acetate buffer at pH 4.5 for 15 minutes at a flow rate of 5 μl/min.
- Ethanolamine is injected for 7 minutes at a flow rate of 10 μl/min to deactivate remaining active esters.
The entire coupling procedure should result in ~ 3000 RU MMTS-papain immobilized in flow cell 2.
Covalent attachment of MMTS-papain for kinetic analysis
The same procedure is used for immobilizing MMTS-papain for kinetic analysis, with the only difference being that in kinetic analysis, the immobilization level needs to be low in order to avoid the binding rate becoming limited by diffusion. Flow cell 1 is left unmodified in this step as well in order to be used as a reference surface.
- After activation of the surface with the mixture of NHS and EDC as in 7.1, MMTS-papain is injected at 1μg/ml for 50 seconds. After that the surface is de-activated with an injection of ethanolamine as in step 7.3.
The coupling level should be approximately 50 RU after this procedure.
7. Determining Cystatin B concentration using the CFCA assay
- The CFCA experiment is set up according to the CFCA wizard in Biacore X100 Plus Package. Approximately 10 nM protein is injected at flow rates 10 and 100 μl/min in duplicate for 48 seconds. The surface is regenerated with 20 mM NaOH between each cycle.
- Include blank injections for each flow rate.
- The protein concentration is then determined from the binding data (Figure 3) using the CFCA evaluation feature of Biacore X100 Plus Package evaluation software.
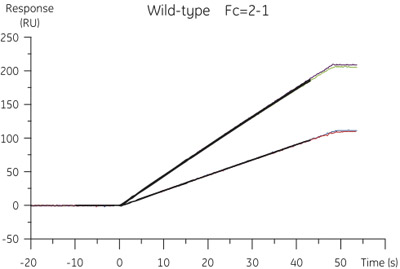
Figure 2. Results from the CFCA analysis of wild-type cystatin B. The specific binding concentration data was extracted from the sensorgrams obtained at flow rates of 10 and 100 μl/min.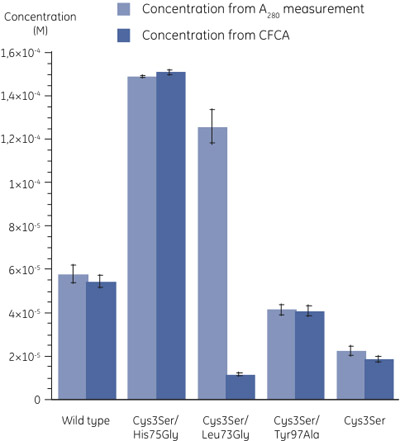
Figure 3. Concentrations of the mutants determined using A280 measurement (n=3) and CFCA (n=2). The standard errors are denoted by error bars. The molar absorption coefficients were calculated as described in (6). A large difference in the concentration values of Cys3Ser/Leu73Gly mutant determined by the two methods can be observed. - When comparing the concentration determined by A280 measurement with that by CFCA, it is clear that, while in most cases the protein is fully active, for the Cys3Ser/Leu73Gly variant, the fraction of active protein is very low (Figure 3). The table below shows the activities of cystatin B variants related to the total concentrations by A280.
Sample CFCA in relation to A280 (%) Wild type 94 Cys3Ser/His75Gly 101 Cys3Ser/Leu73Gly 9 Cys3Ser/Tyr97Ala 99 Cys3Ser 83
8. Measuring the Kinetics of Cystatin B binding to Papain
- Based on the CFCA measurements, prepare a two-fold concentration series ranging from 2.5 to 40 nM for each of the cystatin B variants.
- The kinetics experiment is set up using the kinetics wizard in Biacore X100 with the single cycle approach. The surface is not regenerated between injections, but after the end of each analysis cycle using 20 mM NaOH as regeneration solution.
- Set up the experiment so that each cycle containing sample is flanked by a blank cycle, where buffer is injected instead of sample.
- Use the kinetics evaluation feature in Biacore X100 to automatically perform blank subtractions of reference subtracted data before fitting a 1:1 interaction model.
- The experimental data obtained for the binding of cystatin B variants to papain is shown in Figure 4. The table below summarizes the association and dissociation rate constants and the equilibrium dissociation constants obtained in the kinetic analysis.
Sample ka (M-1 s-1) kd (s-1) KD (M) Wild type 1.8 x 106 0.41 x 10-3 2.3 x 10-10 Cys3Ser/His75Gly 1.1 x 106 1.7 x 10-3 1.5 x 10-9 Cys3Ser/Leu73Gly 1.1 x 106 23 x 10-3 2.2 x 10-8 Cys3Ser/Tyr97Ala 1.7 x 106 12 x 10-3 7.1 x 10-9 Cys3Ser 0.9 x 106 0.53 x 10-3 5.8 x 10-10 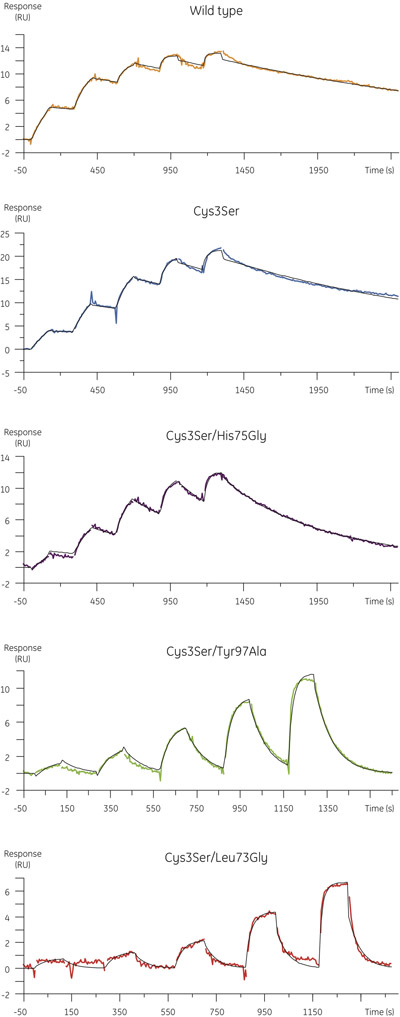
Figure 4. Kinetic profiles for cystatin B variants binding to MMTS-papain determined using single cycle kinetics. Sensorgrams show blank- and reference subtracted data with kinetic fit for 1:1 interaction model overlaid in black. - While the association rates of the mutants are comparable to that of the wild type protein (Figure 5, upper panel), the dissociation rate constants of the mutants can be higher by close to two orders of magnitude, with a corresponding increase in the equilibrium dissociation constant (Figure 5, lower panel).

Figure 5. The calculations of the rate and affinity constants were based on the concentrations obtained from CFCA. The changes of ka, kd and KD, associated with the mutations are depicted in the graphs. - Since sample concentration is a parameter in the calculation of the association rate constant from the experimental data, it is important that it is correct. Using the concentration based on the A280 measurement instead of CFCA would lead to the conclusion that Leu73 is important for the association rate, where replacement seems to result in about 10 times slower association than the other cystatin B variants. However, the specific binding concentration measured by CFCA is only about 10% of the total protein in this case. When this is taken this into account it becomes clear that the association rate for Leu73 is similar to that of the other variants. Therefore, CFCA allows for correct assessment of ka and KD thus making possible the appropriate interpretation of the interaction mechanism.
Dyskusje
In this work, four mutants and wild type cystatin B were produced in order to assess the importance of the second binding loop and C-terminal ends for the interaction between cystatin B and papain. This study demonstrated the advantage and ease of using Biacore X100 to determine specific binding concentration and to analyze the kinetics of protein-protein interactions in order to understand structure-function relationships. We showed that measuring total protein concentration does not reveal the protein variants that have reduced binding activity, which in this case introduces a significant measurement error in the determination of binding affinity and kinetics. The specific binding concentrations of the cystatin B variants were determined using CFCA with Biacore X100. Using concentration measurements by CFCA as input to the kinetic analysis resulted in reliable rate and affinity constants, thus allowing the right interpretation of the interaction mechanism.
The decreased affinities of the mutants were almost exclusively due to an increased kd-value, whereas ka was affected only slightly. This behavior indicates that both the second binding loop region and the C-terminal end are not important for the binding rate of inhibitor to papain. Instead, they contribute to binding affinity primarily by keeping the inhibitor attached to the enzyme once the complex has been formed.
Ujawnienia
The author is employed by GE Healthcare that produces reagents and instruments used in this article.
Materiały
| Name | Company | Catalog Number | Comments |
| Biacore™ X100 System | GE Healthcare | BR-1100-73 | http://www.biacore.com/lifesciences/products/systems_overview/x100/system_information/index.html |
| Biacore™ X100 Plus Package | GE Healthcare | BR-1007-98 | http://www.biacore.com/lifesciences/products/systems_overview/x100/system_information/index.html |
| Sensor Chip CM5 | GE Healthcare | BR-1000-14 | http://www.biacore.com/lifesciences/products/systems_overview/x100/system_information/index.html |
| Amine Coupling Kit | GE Healthcare | BR-1000-50 | |
| Acetate buffer pH 4.5, 50 ml | GE Healthcare | BR-1003-50 | |
| HBS-EP+ buffer 10X, 4 x 50 ml | GE Healthcare | BR-1008-26 | |
| Plastic Vials 11 mm | GE Healthcare | BR-1002-87 | |
| Rubber caps, type 2 | GE Healthcare | BR-1004-11 |
Odniesienia
- Christensen, L. L. H. Theoretical analysis of protein concentration determination using biosensor technology under conditions of partial mass transport limitation. Anal. Biochem. 249, 153-164 (1997).
- Sigmundsson, K., Mâsson, G., Rice, R., Beauchemin, N., Öbrink, B. Determination of active concentrations and association and dissociation rate constants of interacting biomolecules: an analytical solution to the theory for kinetic and mass transport limitations in biosensor technology and its experimental verification. Biochemistry. 41 (26), 8263-8276 (2002).
- Stubbs, M. T., Laber, B., Bode, W., Huber, R., Jerala, R., Lenarcic, B., Turk, V. The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 9 (6), 1939-1947 (1990).
- Pol, E., Björk, I. Importance of the second binding loop and the C-terminal end of cystatin B (Stefin B) for inhibition of cysteine proteinases. Biochemistry. 38 (32), 10519-10526 (1999).
- Björk, I., Pol, E., Raub-Segall, E., Abrahamson, M., Rowan, A. D., Mort, J. S. Differential changes in the association and dissociation rate constants for binding of cystatins to target proteinases occurring on N-terminal truncation of the inhibitors indicate that the interaction mechanism varies with different enzymes. Biochem. J. 299, 219-225 (1994).
- Pace, C. N., Vajdos, F., Fee, L., Grimsley, G., Gray, T. How to measure and predict the molar absorption coefficient of a protein. ProteinScience. 4 (11), 2411-2423 (1995).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone