Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Identification of Growth Inhibition Phenotypes Induced by Expression of Bacterial Type III Effectors in Yeast
W tym Artykule
Podsumowanie
In this video, we describe a procedure for the expression of bacterial type III effectors in yeast and the identification of effector-induced growth inhibition phenotypes. Such phenotypes can be subsequently exploited to elucidate effector functions and targets.
Streszczenie
Protokół
I. Designing a Yeast Expression System for Type III Effectors
Calibrating a yeast system appropriate for expression of the type III effector(s) of interest is an important task and may require some trial and error. Factors of major relevance that should be considered and optimized when designing such a system are: 1) the promoter driving expression of the effector(s), 2) the copy number of the effector gene, 3) the epitope tag used to verify protein expression, and 4) the yeast strain.
1) Promoter
Because bacterial type III effectors may be toxic to yeast cells, an inducible promoter should be used to control their expression. The GAL1 promoter is commonly used for this purpose and its activity is regulated by the carbon source present in the growth medium: it is repressed by glucose and induced by galactose. However, this promoter was reported as slightly leaky under repressing conditions resulting in undesired toxicity. If a similar problem is encountered, it is advisable to minimize the copy number of the effector gene as described below, or to use alternative inducible promoters, such as the MET3 or CUP1 promoters1. Here we describe procedures for expressing effector proteins from a galactose-inducible promoter.
2) Gene copy-number
An additional parameter affecting the level of protein expression is the number of copies of the effector gene present in the cell. High expression levels are achieved when the effector gene is carried by a 2-micron plasmid (40-60 copies per cell). Intermediate expression is obtained by using a centromere-containing plasmid (1-3 copies per cell), whereas low expression is achieved when the effector gene is integrated into the yeast genome by homologous recombination. By combining a centromere-containing vector and the GAL1 promoter we have successfully expressed about 50 effectors of the plant pathogens Xanthomonas campestris pv. vesicatoria and Pseudomonas syringae pv. tomato at levels detectable by immunoblot analysis and with negligible leakage under repressing conditions (Salomon and Sessa, unpublished).
3) Epitope tag
If antibodies against the effector of interest are not available, an epitope tag is fused to the effector protein to monitor its expression by immunoblot. Commonly used tags are Myc, Hemagglutinin (HA) or Flag, to which reliable commercial antibodies are available. We suggest using only one copy of the tag to minimize undesired deleterious effects on structure and activity of the effector protein.
4) Yeast strain
A fundamental requirement for the yeast strain to be used for expression is auxotrophy to the selectable marker of the expression vector. It is important to note that different yeast strains may display different sensitivities to expression of certain effectors. For example, we observed that the BY4741 and W303 strains have different sensitivities to several Xanthomonas campestris pv. vesicatoria effectors (Salomon and Sessa, unpublished). Therefore, it is preferable to test the effector(s) of interest in different yeast strains.
II. Preparation of Yeast Media
For growth of yeast strains that do not contain any vector, use YPD (10 g/L yeast extract, 20 g/L peptone and 2% [w/v] glucose) as the growth medium. For solid media, add 2% [w/v] agar to the solution (if the solid medium is very soft, add 0.05% [v/v] from a 5 N NaOH stock solution to the medium before autoclaving). We recommend preparing the medium without glucose in 90% of the final volume and autoclaving it. Shortly before use, fill the volume to 100% with a filter-sterilized 20% [w/v] glucose solution (keep the 20% glucose solution at 4°C to prevent contamination).
For growth of yeast strains containing a vector that provides prototrophy to a selectable marker (leucine, uracil, histidine or tryptophan), use synthetic drop-out medium without leucine, uracil, histidine and tryptophan (6.7 g/L yeast nitrogen base without amino acids, 1.4 g/L yeast synthetic drop-out medium supplement) and containing 2% [w/v] glucose, or 2% [w/v] galactose and 1% [w/v] raffinose. For solid media, add 2% [w/v] agar to the solution. We recommend preparing the medium in 90% of the final volume and autoclaving it. Shortly before use, fill the volume to 100% with a filter-sterilized 20% glucose solution, or a filter-sterilized 20% galactose + 10% raffinose solution, depending on the desired carbon source (keep the 20% glucose and the 20% galactose + 10% raffinose solutions at 4°C to prevent contamination). Depending on the selectable marker of the expression vector, add the other amino acids or uracil before use (10 ml/L from a 1 g/100 ml leucine stock, 10 ml/L from a 200 mg/100 ml uracil stock, 2 ml/L from a 1 g/100 ml histidine stock, or 2 ml/L from a 1 g/100 ml tryptophan stock). When preparing leucine and uracil stocks, sterilize them by autoclaving and keep at room temperature. When preparing histidine and tryptophan stocks, sterilize them by filtering, wrap the bottle with aluminum foil to protect from light, and keep at 4°C. In the procedures described below, synthetic drop-out medium supplemented with leucine, uracil, histidine and tryptophan is designated as synthetic complete medium.
Solid-media plates can be stored for up to two months at 4°C. Before use, dry the plates for 20 min in a sterile laminar flow hood at room temperature.
III. Yeast Transformation
- Before starting the yeast transformation procedure, prepare the following three solutions: a 50% [w/v] polyethylene glycol (PEG) 3350 solution, a 1 M LiAc solution pH=8.0, and a TE solution (100 mM Tris pH=6.85 and 10 mM EDTA pH=8.0). Filter-sterilize the solutions and keep at 4°C.
- Using a long wooden shaft or a sterile inoculation loop, pick a yeast colony (1-2 mm diameter) from a fresh plate and inoculate 3 ml of YPD in a 15 ml polypropylene tube. Vortex briefly. Place the culture tube in a roller and incubate overnight at 30°C with constant rotation.
- In the morning, remove the culture from the roller and centrifuge at 800 g for 5 min at room temperature. After centrifugation, completely remove the supernatant.
- Resuspend the cells in 1 ml resuspension buffer (10% [v/v] TE solution, 10% [v/v] 1 M LiAc solution) and mix by vortexing.
- For each plasmid to be transformed and an additional control transformation, transfer 100 μl of the cell suspension to a microtube.
- To each tube, add 5 μl of single strand salmon sperm DNA (10 mg/ml) and 250-500 ng of plasmid DNA to be transformed (except for the control tube).
- Carefully add to each tube 650 μl of transformation solution (10% [v/v] TE solution, 10% [v/v] 1 M LiAc solution, 80% [v/v] of 50% [w/v] PEG solution) and vortex.
- Incubate at 30°C for 30 min on a roller.
- Remove the tubes from the shaker, add 70 μl of DMSO, and mix by inverting the tubes 10-15 times (do not vortex to avoid breaking the cells).
- Place the tubes in a 42°C pre-warmed water bath and incubate for 15 min.
- Remove the tubes from the water bath and immediately place them on ice for 2 min.
- Once the tubes have cooled down, centrifuge them at 800 g for 5 min at room temperature.
- Take the tubes out of the microcentrifuge and carefully remove the viscous supernatant using a pipettor.
- Resuspend the cell pellet of each tube in 150 μl of sterile double distilled water (DDW), and spread the cell suspension on a synthetic complete medium plate containing 2% glucose as a carbon source and without the amino acid or nucleotide to which the plasmid provides prototrophy.
- Leave the plates open for 2 min and allow them to dry in a laminar flow hood. Place the plates in a 30°C incubator for two-three days.
- Once colonies (1-2 mm diameter) have appeared on the plates, pick approximately 10 single colonies from each transformation and transfer them to a fresh plate. When transferring the colonies, make 2 cm x 2 cm patches to allow growth of a large amount of yeast cells. Incubate the plates in an incubator at 30°C for two days.
- When the yeast patches have grown, remove the plates from the incubator and seal them with parafilm. Plates can be stored at 4°C. Transfer colonies to a fresh plate every 10-14 days.
IV. Preparing a Yeast Protein Extract to Verify Effector Expression by Immunoblot
- Pick a yeast colony (1-2 mm diameter) from a fresh plate to a 15 ml polypropylene tube containing 3 ml of the appropriate synthetic complete medium supplemented with 2% glucose as a carbon source and without the amino acid or nucleotide to which the plasmid of choice provides prototrophy. Place the culture tube in a roller and incubate overnight at 30°C.
- In the following day, remove the culture from the roller and centrifuge at 800 g for 5 min at room temperature. Remove the tubes from the centrifuge and discard the supernatant.
- Resuspend the cell pellet in 3 ml of sterile DDW and mix by vortexing.
- Centrifuge again, discard supernatant, and resuspend cells in 3 ml of sterile DDW.
- Transfer 200 μl of the cell suspension to a new 15 ml tube containing 3 ml of synthetic complete medium supplemented with 2% galactose and 1% raffinose as a carbon source, and without the amino acid or nucleotide to which the plasmid of choice provides prototrophy. Mix by vortexing, place in a roller, and incubate overnight at 30°C.
- In the following morning, transfer 1 ml of the culture (or more for low density cultures) to a microtube. Harvest the cells by centrifugation at 800 g for 5 min at room temperature.
- From here on, work in a fume hood to avoid breathing β-mercaptoethanol toxic vapors. Carefully remove the supernatant using a pipettor and resuspend the cell pellet in 100 μl of ice-cold lysis solution (4% [v/v] 5 N NaOH, 0.5% [v/v] β-mercaptoethanol). Mix vigorously by vortexing and incubate on ice for 30 min.
- During the incubation, determine the volume of HCl (6 N) required for titering 100 μl of lysis buffer to pH 9-10. To this aim, place 10 microtubes in a rack and transfer 100 μl of the lysis buffer into each of them. Add various volumes (1-10 μl) of HCl (6 N) to each tube and mix by vortexing. Take a sample and check the pH of the solution using a pH indicator strip.
- At the end of the incubation, add the HCl volume determined in the previous step to the lysate and mix by vortexing (for better accuracy, place the HCl solution on the side of the tube without inserting the tip into the lysate).
- Add 50 μl of 3x sample buffer (30% [v/v] glycerol, 15% [v/v] β-mercaptoethanol, 37.5% [v/v] 500 mM Tris-HCl pH=6.8, 0.15% [w/v] sodium dodecyl sulfate [SDS] and few grains of bromophenol blue) to the lysate and mix by vortexing (if the solution turns yellow, it means that the pH is too low, and a few microliters of lysis solution should be added until the solution turns blue).
- Boil the lysate solution for 5 min on a heating block after puncturing a hole in the cover of the microtube with a needle.
- Remove the microtube from the heating block and allow the solution to cool for 2 min at room temperature.
- Vortex the lysate solution and then spin it down for 10 sec. Load 30 μl onto a SDS-PAGE gel for immunoblot analysis.
V. Spotting Assay to Detect Effector-induced Growth Inhibition Phenotypes
- From a fresh plate pick a yeast colony (1-2 mm diameter) carrying the expression plasmid of interest and inoculate, in a 15 ml polypropylene tube, 3 ml of synthetic complete medium supplemented with 2% glucose as a carbon source and without the amino acid or nucleotide to which the expression plasmid provides prototrophy. Repeat the same procedure for a control yeast strain carrying an empty plasmid. Place the culture tubes in a roller and incubate overnight at 30°C with constant rotation.
- In the following day, remove the cultures from the roller and centrifuge at 800 g for 5 min at room temperature. Remove the tubes from the centrifuge and discard the supernatant.
- Resuspend the cell pellet in 3 ml of sterile DDW and mix by vortexing.
- Repeat the centrifugation step and resuspend cells in 3 ml of sterile DDW.
- To determine their optical density (OD), transfer 100 μl of each culture to a microtube containing 900 μl DDW and vortex. Pour the content of the tubes into a plastic cuvette and measure the absorbance at a wavelength of 600 nm (use as reference a cuvette filled with 1 ml of DDW). Calculate the OD600 of the initial cultures by multiplying the OD600 of the cultures in the cuvette by a dilution factor of 10.
- Next, based on the OD600 of the initial cultures, prepare cell suspensions (1 ml) with an OD600=1.0 in sterile microtubes.
- By using 3 sterile microtubes filled with 900 μl sterile DDW, prepare three 10-fold serial dilutions (OD600=0.1, 0.01 and 0.001) from each cell suspension (OD600=1.0) prepared in the previous step.
- Prepare two plates containing synthetic complete medium without the amino acid or nucleotide to which the plasmid of choice provides prototrophy. The medium of one plate should be supplemented with 2% glucose and that of the other plate with 2% galactose and 1% raffinose. Dry the plates in a sterile laminar flow hood at room temperature for 20 min and place them on a grid.
- For each culture, spot 10 μl from the four dilutions in a row on both the repressing and inducing media.
- After spotting, leave the plates open in the hood for several minutes. When the spots are dry, cover the plates and place them in a 30°C incubator for 2-3 days.
- After incubation, take out the plates and analyze yeast growth. First, confirm that the cell densities are similar on the plate containing repressing medium. Then, compare the growth of the culture expressing the effector protein of interest with the culture carrying an empty vector on the plate containing inducing medium.
Note: effectors may target cellular processes that are not rate-limiting to yeast growth under standard laboratory growth conditions. To allow the identification of growth inhibition phenotypes for such effectors, inducing media can be supplemented with compounds that alter yeast cellular functions, thus increasing their sensitivity to effectors. For a list of chemicals that can possibly be integrated into this procedure, refer to Parsons et al. (2004)2.
VI. Representative Results
A representative yeast spotting assay and detection of growth inhibition phenotypes induced by expression of type III effectors are shown in Fig. 1. In this experiment, the type III effectors AvrPto, HopAA1-1 and AvrE1 of the Gram-negative phytopathogenic bacterium Pseudomonas syringae pv. tomato (Pst) were expressed from the centromere-containing plasmid pGML10 in the yeast strain BY4741, and tested for their ability to inhibit yeast growth. Individual yeast cultures expressing Pst type III effectors under the control of the galactose inducible GAL1 promoter or containing an empty vector were serially diluted and plated onto repressing (glucose) or inducing (galactose) media (Fig. 1). On repressing medium, yeast strains carrying plasmids for the expression of AvrPto and HopAA1-1 exhibited similar growth as the control strain containing an empty vector. However on the same medium, yeast carrying AvrE1 displayed a slightly reduced growth, probably related to some degree of leakage of the GAL1 promoter and to the high cytotoxic effect of AvrE1. In inducing conditions, expression of the AvrE1 effector caused a drastic growth inhibition phenotype reflected by the lack of colonies in any dilution. As previously observed by Munkvold et al.3, expression of HopAA1-1 also resulted in severe inhibition of growth, while AvrPto did not show any effect.
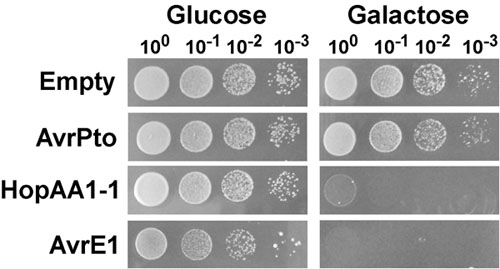
Figure 1. Yeast growth inhibition caused by expression of the Pseudomonas syringae pv. tomato (Pst) type III effectors AvrE1 and HopAA1-1. Yeast strains (BY4741) containing the plasmid pGML10, either empty or carrying GAL1-driven cassettes for galactose-inducible expression of AvrPto, HopAA1-1 or AvrE1, were grown overnight in synthetic complete medium supplemented with glucose (2%) as a carbon source, and lacking leucine. Cultures were washed, normalized to OD600=1.0 and serial 10-fold dilutions were spotted onto synthetic complete solid media lacking leucine and containing glucose (2%), or galactose (2%) and raffinose (1%). Photographs were taken after 2 and 3 days of growth at 30°C for yeast growing in glucose and galactose media, respectively.
Dyskusje
In this presentation, we illustrated how to use the budding yeast Saccharomyces cerevisiae as a heterologous system for the expression of type III bacterial effector proteins and how to identify effector-induced growth inhibition phenotypes. Importantly, these phenotypes can be utilized in genetic screens to identify suppressors of the negative impact of effectors on yeast growth. Suppressors may represent either direct targets of the effector studied or proteins that participate in cellular processes affected b...
Podziękowania
This work was supported by the Israel Science Foundation.
Materiały
| Name | Company | Catalog Number | Comments |
| Yeast extract | Difco Laboratories | 212750 | |
| Peptone | Difco Laboratories | 211677 | |
| D-glucose | Sigma-Aldrich | G5767 | |
| Agar | Difco Laboratories | 214010 | |
| Sodium hydroxide (NaOH) | Sigma-Aldrich | S8045 | |
| Yeast nitrogen base w/o amino acids | Difco Laboratories | 291940 | |
| Yeast synthetic drop-out medium supplement | Sigma-Aldrich | Y2001 | |
| D-galactose | Sigma-Aldrich | G0750 | >99%; <0.1% glucose |
| D-raffinose | Sigma-Aldrich | R0250 | >98% |
| L-leucine | Sigma-Aldrich | L8000 | |
| Uracil | Sigma-Aldrich | U0750 | |
| L-tryptophan | Sigma-Aldrich | T0254 | |
| L-histidine | Sigma-Aldrich | H6034 | |
| DNA, single stranded, from salmon testes | Sigma-Aldrich | D7656 | |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D5879 | Desiccate |
| Hydrochloric acid (HCl) | Sigma-Aldrich | H1758 | |
| Polyethylene glycol (PEG) 3350 | Sigma-Aldrich | P4338 | |
| Lithium acetate (LiAc) | Sigma-Aldrich | L4958 | |
| Tris (base) | JT Baker | 4109-02 | |
| Ethylenediamine-tetraacetic acid (EDTA) | Sigma-Aldrich | E5134 | |
| β-mercapt–thanol | Sigma-Aldrich | M6250 | |
| Glycerol | Sigma-Aldrich | G5516 | |
| Bromophenol blue | Sigma-Aldrich | B6131 | |
| Dodecyl sulfate sodium salt (SDS) | Merck & Co., Inc. | 8.22050.1000 | |
| Centrifuge tubes (15 ml) | Corning | 430052 | Sterile |
| Spectrophotometer cuvette (10x4x45 mm) | Sarstedt Ltd | 67.742 | |
| Inoculation loop | Sigma-Aldrich | Z643009 | Sterile |
| Parafilm | Sigma-Aldrich | P7543 | |
| pH indicator strip, pH 6.5-10.0 | Merck & Co., Inc. | 1.09543.0001 |
Odniesienia
- Siggers, K. A., Lesser, C. F. The yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe. 4, 8-15 (2008).
- Parsons, A. B. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22, 62-69 (2004).
- Munkvold, K. R., Martin, M. E., Bronstein, P. A., Collmer, A. A survey of the Pseudomonas syringae pv. tomato DC3000 type III secretion system effector repertoire reveals several effectors that are deleterious when expressed in Saccharomyces cerevisiae. Mol. Plant-Microbe Interact. 21, 490-502 (2008).
- Curak, J., Rohde, J., Stagljar, I. Yeast as a tool to study bacterial effectors. Curr. Opin. Microbiol. 12, 18-23 (2009).
- Slagowski, N. L., Kramer, R. W., Morrison, M. F., LaBaer, J., Lesser, C. F. A functional genomic yeast screen to identify pathogenic bacterial proteins. PLoS Pathog. 4, e9-e9 (2008).
- Huang, J., Lesser, C. F., Lory, S. The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature. 456, 112-115 (2008).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone