Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Constant Pressure-controlled Extrusion Method for the Preparation of Nano-sized Lipid Vesicles
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
This protocol describes an extrusion method for preparing lipid vesicles of sub-micron sizes with a high degree of homogeneity. This method uses a pressure-controlled system with controlled nitrogen flow rates for liposome preparation. The lipid preparation1,2, liposome extrusion, and size characterization will be presented herein.
Streszczenie
Liposomes are artificially prepared vesicles consisting of natural and synthetic phospholipids that are widely used as a cell membrane mimicking platform to study protein-protein and protein-lipid interactions3, monitor drug delivery4,5, and encapsulation4. Phospholipids naturally create curved lipid bilayers, distinguishing itself from a micelle.6 Liposomes are traditionally classified by size and number of bilayers, i.e. large unilamellar vesicles (LUVs), small unilamellar vesicles (SUVs) and multilamellar vesicles (MLVs)7. In particular, the preparation of homogeneous liposomes of various sizes is important for studying membrane curvature that plays a vital role in cell signaling, endo- and exocytosis, membrane fusion, and protein trafficking8. Several groups analyze how proteins are used to modulate processes that involve membrane curvature and thus prepare liposomes of diameters <100 - 400 nm to study their behavior on cell functions3. Others focus on liposome-drug encapsulation, studying liposomes as vehicles to carry and deliver a drug of interest9. Drug encapsulation can be achieved as reported during liposome formation9. Our extrusion step should not affect the encapsulated drug for two reasons, i.e. (1) drug encapsulation should be achieved prior to this step and (2) liposomes should retain their natural biophysical stability, securely carrying the drug in the aqueous core. These research goals further suggest the need for an optimized method to design stable sub-micron lipid vesicles.
Nonetheless, the current liposome preparation technologies (sonication10, freeze-and-thaw10, sedimentation) do not allow preparation of liposomes with highly curved surface (i.e. diameter <100 nm) with high consistency and efficiency10,5, which limits the biophysical studies of an emerging field of membrane curvature sensing. Herein, we present a robust preparation method for a variety of biologically relevant liposomes.
Manual extrusion using gas-tight syringes and polycarbonate membranes10,5 is a common practice but heterogeneity is often observed when using pore sizes <100 nm due to due to variability of manual pressure applied. We employed a constant pressure-controlled extrusion apparatus to prepare synthetic liposomes whose diameters range between 30 and 400 nm. Dynamic light scattering (DLS)10, electron microscopy11 and nanoparticle tracking analysis (NTA)12 were used to quantify the liposome sizes as described in our protocol, with commercial polystyrene (PS) beads used as a calibration standard. A near linear correlation was observed between the employed pore sizes and the experimentally determined liposomes, indicating high fidelity of our pressure-controlled liposome preparation method. Further, we have shown that this lipid vesicle preparation method is generally applicable, independent of various liposome sizes. Lastly, we have also demonstrated in a time course study that these prepared liposomes were stable for up to 16 hours. A representative nano-sized liposome preparation protocol is demonstrated below.
Protokół
1. Liposome Preparation
- Retrieve a 20 mL glass vial with a Teflon-lined cap.
- Clean all glassware and syringes with chloroform prior to use to prevent contamination.
- Transfer 100 μL of reagent grade chloroform to the glass vial using a 250 μL air-tight glass syringe.
- Add 30 μL of reagent grade methanol to the same glass vial using a 100 μL air-tight glass syringe.
- To prepare a 2 mM 7:1.5:1.5 phosphatidylcholine (POPC): phosphatidylethanolamine (POPE): cholesterol lipid solution, transfer 216 μL of 10 mg/ mL POPC, 42 μL of 10 mg/mL POPE and 20 μL of 11 mg/mL cholesterol solutions in CHCl3 to the 20 mL glass vial.
- Evaporate the organic solvents using slow-flow argon or nitrogen gas until a thin film of lipids is observed on the bottom of the vial.
- Place the uncapped glass vial in a vacuum desiccator for at least 30 minutes to remove residual solvent.
- Transfer 2 mL of buffer, previously passed through a 0.2 micron filter, to the glass vial to hydrate the lipids.
- Incubate the mixture at 4 °C overnight and use within 48 hours.
2. Freeze-and-thaw: For Liposome Sizes of 30 - 100 nm Only
- Freeze liposome suspension in liquid nitrogen for 15 seconds.
- Thaw liposome suspension using a heating block at 42 °C temperature for ~3 minutes.
- Repeat steps 2.1 and 2.2 for a total of 5 cycles.
3. Extrusion
Follow the Avestin instructions to correctly assemble the Liposofast LF-50 extruder, using the instrument outline in their guide book.
- Place the large hole support screen (with circular holes) in the support filter base, followed by the circular sintered dish (without holes), one drain disc (25 mm in diameter), and one polycarbonate membrane (25 mm in diameter).
- Place the small, black O-ring on top of the membrane to secure it to the support filter base.
- Attach the top filter extruder by tightening 4 screws in the 4 corresponding holes.
- Assemble the filter extruder unit to the underneath the large extruder barrel. Add the liposome solution to the cylinder barrel. Place the following on top of the extrusion barrel in order: large circular O-ring, narrow cap, large circular O-ring, circular cap.
- Attach the gas regulator to the extruder barrel top and close all valves to prevent air leakage, including the pressure relief valve. Place a 20 mL glass vial or 50 mL Erlenmeyer flask under the extruder filter unit. A safety valve is connected to the regulator and will release if pressure exceeds 600 psi. Turn on the nitrogen gas and open the gas valve connected to the extruder.
- Increase nitrogen pressure to 25 psi for 400 nm liposomes, 125 psi for 100 nm liposomes, and 400-500 psi for 30 nm liposomes.
- Watch the liposome suspension as it ejects into the container while being pushed by the nitrogen. Keep the nitrogen flowing until you no longer observe any liquid passing through the extruder filter into the glass vial.
4. Dynamic Light Scattering (DLS) Analysis
- Prepare 50 μL of a 20 μM liposome solution.
- Turn on the power source and the lamp source.
- Open the DynaPro software.
- Set the software to the MS/X algorithm model to detect liposomes of 30 nm and 100 nm and MS800 algorithm model to detect >100 nm liposomes.
- Connect to the hardware.
- Pipet 14 μL of liposome sample into the quartz cuvette and insert into the cell holder.
- Press start.
- Press stop after ~20 - 30 acquisitions.
- Analyze the average liposome diameter peaks recorded on the histogram.
5. Nanoparticle Tracking Analysis (NTA)
- Prepare a 500 μL, 0.1 μM liposome solution.
- Rinse the sample compartment with water and ethanol.
- Dry the sample compartment with a lint-free paper towel.
- Turn on the laser power source and the computer.
- Deliver 300 μL of 0.1 μM liposome solution to the sample compartment.
- Open the Temperature control and the Nanoparticle Tracking Analysis software.
- Press Capture button to turn on the laser.
- Use the horizontal and vertical adjustments to move the stage and adjust the microscope focus.
- Set the desired temperature (20 °C) and recording time (at least 30 seconds).
- Press the Record button to take multiple picture frames of the liposome particles for a specified amount of time. Analyze the peaks corresponding to the diameter liposome sizes on the histogram as it tracks the motion of each particle.
6. Representative Results
A scheme outlining the extrusion method is presented in Figure 1. To obtain optimum results, preparation of liposomes with diameters of 30 nm requires a high pressure of ~ 500 psi and diameters of 100 nm requires a pressure of 125 psi to achieve a rapid filter rate. For diameters of 400 nm, a low pressure of ~ 25 psi is recommended to achieve a slower filter rate, which allows the vesicles to elongate and form larger, homogeneous liposomes. We conducted a series of experiments to determine the optimal pressure for producing consistent sub-micron vesicle sizes. We varied the pressure as well as the number of extrusion passes through polycarbonate filters with pore sizes of 30, 100, and 400 nm and discovered appropriate pressure for each desired size. For 30 nm pores, pressure below 500 psi will reduce the flow rate, causing elongation and thus larger vesicle sizes. For 100 nm pores, a steady flow was achieved at 125 psi. For 400 nm pores, low pressure (25 psi) allows the vesicles to elongate into larger vesicle sizes. A slow drop-wise filtration flow is optimal to create larger sub-micron vesicles13.
We performed DLS to determine the liposome sizes extruded through three different diameter sizes, i.e. 30, 100 and 400 nm. DLS is an established method that collects scattered light to determine the particle diameter. We extruded hydrated liposomes of 2 mM through a polycarbonate 30 nm membrane at 500 psi with 5 passes through the filter pore; a 100 nm polycarbonate membrane at 125 psi with 5 passes through the filter pore and a 400 nm polycarbonate membrane at 25 psi with 2 passes through the filter pore. The diameters of the liposomes and measured by DLS for 30, 100 and 400 nm pore sizes were 66 ± 28, 138 ±18 and 360 ± 25 nm (Figure 2). A suspension of 50 nm polystyrene beads was used as a calibration standard as shown in Figure 2, where it recorded a diameter of 47 ± 16 nm. The percent polydispersity shows that there is no overlap within liposome sizes. It is typical to observe diameters higher than 30 nm when using DLS analysis due to the known bias this instrument has towards larger particles12. Light scatter intensities from large and small particles are collected simultaneously from one detection method and thus harder to resolve liposomes in suspension12. Despite this instrumental limitation, the calibration curve describes a near linear correlation.
NTA is a new technology that measures the size of each particle from direct observations of diffusion in a liquid medium, independent of particle refractive index or density. This high-resolution technique can be used to supplement the measurement of liposomes with DLS. The NTA recorded diameters of 95 ± 48 and 356 ± 51 nm, two 100 nm and 400 nm polystyrene solutions were used for calibration. Lipid solutions of 30 nm and 100 nm were observed to have a more comparable diameter relative to the DLS as shown in Figure 3, producing average sizes of 29 ± 14, 95 ± 17 and 359 ± 73 nm. The NTA may be a more general characterization technique to quantify microscopic particles since its sensitivity allows it to measure particles of 50 - 1000 nm. The calibration curve shows a linear correlation between the polycarbonate membrane pores versus the recorded NTA diameter.
| Liposome Sizes | Detection Threshold | Expected Minimum Particle Size (nm) |
| 30 nm | 11 | 30 |
| 100 nm | 11 | 100 |
| 400 nm | 21 | 400 |
Table 1. Nanosight Parameters.

Figure 1. Flow chart of the extrusion method, describing how the pressure and nitrogen flow controls the homogeneity of different liposome diameters. Following liposome hydration, unilamellar vesicles of different sizes are extruded through different polycarbonate membrane filter pores at different pressures.
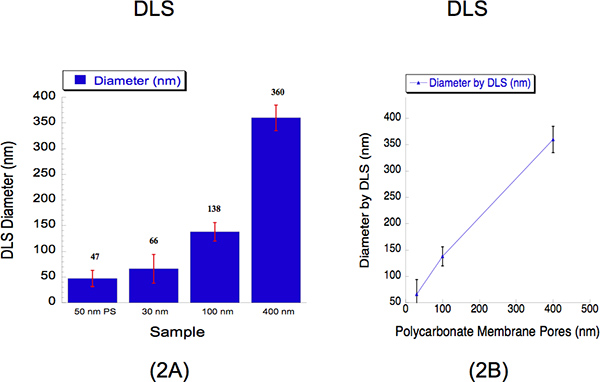
Figure 2. Dynamic Light Scattering (DLS) data describing the quantitative liposome sizes following extrusion. (a) Bar graphs are shown to describe the diameters of the three liposome samples. A calibration standard of 50 nm polystyrene (PS) beads was used as a reference. The average liposome diameters are indicated above the bar for each sample. The x-axis describes the pore sizes of which the solutions were extruded through. The y-axis describes the diameter recorded by DLS. Although the 30 nm and 100 nm sizes recorded values higher than their pore sizes while the larger 400 nm size recorded a slightly lower size, the calibration curve (b) shows a near linear correlation, where the x-axis expresses the polycarbonate membrane pore sizes, and the y-axis describes the recorded liposome diameter by DLS.

Figure 3. Nanoparticle Tracking Analysis (NTA) data describing the liposome sizes following extrusion. (a) The bar graphs represent the diameter of each liposome sample. The x-axis describes the pore sizes of which the solutions were extruded through. The y-axis describes the size diameter recorded by the NTA. The average liposome diameters are labeled above each sample. (b) The NTA calibration curve shows a more linear correlation than the DLS calibration curve between the filter membrane pore sizes versus the recorded diameter. The x-axis describes the polycarbonate membrane pore sizes. The y-axis describes the recorded liposome diameter by NTA.
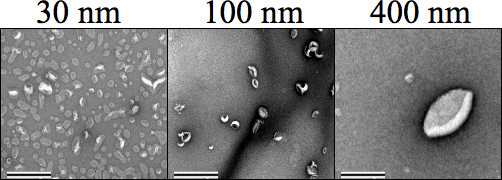
Figure 4. Negative stain transmission electron microscopy (TEM) images showing each liposome size (500 μM) followed by extrusion. Carbon Formvar mesh grids were negatively discharged prior to sample staining, where 1% Uranyl acetate in water was used to stain the samples prior to drying and imaging at 34,000× magnification. Sizes 30, 100, and 400 nm are clearly distinct from each other. The magnification was set to 25,000×. The scale bar represents 0.5 μm.
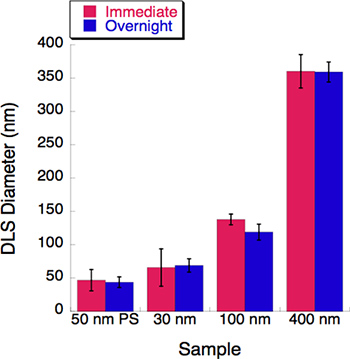
Figure 5. A time-course experiment was conducted with three liposome sizes. Dynamic light scattering (DLS) was measured for each liposome solution immediately following extrusion. All liposome solutions were stored in 4 °C overnight. Their diameters were recorded by the DLS after an overnight incubation. Little to no change was observed after a 16-hour incubation period.
Access restricted. Please log in or start a trial to view this content.
Dyskusje
Using the Avestin Liposofast LF-50 Extruder, we demonstrated how small-sized, synthetic liposomes are prepared through a pressure-controlled system. It is important to note that multilamellar vesicles form spontaneously following liposome hydration, which may lead to production of smaller nanoparticles. These small multilamellar vesicles will inevitably flow through the larger polycarbonate membrane pore size, causing heterogeneity in solutions of unilamellar vesicles produced by a large filter pore. Therefore, it is rec...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
No conflicts of interest declared.
Podziękowania
This work was supported by the Howard Hughes Medical Institute (HHMI) Collaborative Innovation Award. L. A. M. was supported by the Signaling and Cellular Regulation National Institute of Health training grant (T32 GM008759) and the NIH Ruth L. Kirschstein Pre-Doctoral Fellow (CA165349-01). We would like to thank Prof. Michael Stowell (CU Boulder), Prof. Douglas Rees and Prof. Rob Phillips (Caltech) for their invaluable comments.
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments |
| Chloroform | Sigma-Aldrich | 02432-25ML | 95% stabilizers |
| High Grade Methanol | Sigma-Aldrich | 179337-4L | |
| Liposofast LF-50 Extruder | Avestin, Inc. | ||
| Phospholipids | Avanti Polar Lipids | ||
| Polycarbonate Pores | Avestin, Inc. | 25 mm diameter | |
| Drain discs PE | Avestin, Inc. | 230600 | 25 mm diameter |
Odniesienia
- Connor, J., Bucana, C., Fidler, I. J., Schroit, A. J. Differentiation-dependent expression of phosphatidylserine in mammalian plasma membranes: quantitative assessment of outer-leaflet lipid by prothrombinase complex formation. Proc. Natl. Acad. Sci. U.S.A. 86, 3184-3188 (1989).
- Smith, S. A., Morrissey, J. H. Rapid and efficient incorporation of tissue factor into liposomes. J. Thromb. Haemost. 2, 1155-1162 (2004).
- Hui, E., Johnson, C. P., Yao, J., Dunning, F. M., Chapman, E. R. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 138, 709-721 (2009).
- Loughrey, H. C., Choi, L. S., Cullis, P. R., Bally, M. B. Optimized procedures for the coupling of proteins to liposomes. J. Immunol. Methods. 132, 25-35 (1990).
- Mui, B., Chow, L., Hope, M. J. Extrusion technique to generate liposomes of defined size. Methods Enzymol. 367, 3-14 (2003).
- Gruner, S. M., Cullis, P. R., Hope, M. J., Tilcock, C. P. Lipid polymorphism: the molecular basis of nonbilayer phases. Annu. Rev. Biophys. Biophys. Chem. 14, 211-238 (1985).
- Guven, A., Ortiz, M., Constanti, M., O'Sullivan, C. K. Rapid and efficient method for the size separation of homogeneous fluorescein-encapsulating liposomes. J. Liposome. Res. 19, 148-154 (2009).
- Zimmerberg, J., Kozlov, M. How proteins produce cellular membrane curvature. Nature Reviews Molecular Cell Biology. 7, 9-19 (2006).
- Chonn, A., Cullis, P. R. Recent advances in liposomal drug-delivery systems. Curr. Opin. Biotechnol. 6, 698-708 (1995).
- Mayer, L. D., Hope, M. J., Cullis, P. R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta. 858, 161-168 (1986).
- Matsuoka, K., Schekman, R. The use of liposomes to study COPII- and COPI-coated vesicle formation and membrane protein sorting. Methods. 20, 417-428 (2001).
- Dragovic, R. A., Gardiner, C., Brooks, A. S., Tannetta, D. S., Ferguson, D., Hole, P., Carr, B., Redman, C., Harris, A. L., Dobson, P. J., Harrison, P., Sargent, I. L. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. , (2011).
- Hunter, D. G., Frisken, B. J. Effect of extrusion pressure and lipid properties on the size and polydispersity of lipid vesicles. Biophys J. 74, 2996-3002 (1998).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone