Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Genetic Manipulation of Cerebellar Granule Neurons In Vitro and In Vivo to Study Neuronal Morphology and Migration
W tym Artykule
Podsumowanie
Neuronal morphogenesis and migration are crucial events underlying proper brain development. Here, we describe methods to genetically manipulate cultured cerebellar granule neurons and the developing cerebellum for the assessment of morphology and migratory characteristics of neurons.
Streszczenie
Developmental events in the brain including neuronal morphogenesis and migration are highly orchestrated processes. In vitro and in vivo analyses allow for an in-depth characterization to identify pathways involved in these events. Cerebellar granule neurons (CGNs) that are derived from the developing cerebellum are an ideal model system that allows for morphological analyses. Here, we describe a method of how to genetically manipulate CGNs and how to study axono- and dendritogenesis of individual neurons. With this method the effects of RNA interference, overexpression or small molecules can be compared to control neurons. In addition, the rodent cerebellar cortex is an easily accessible in vivo system owing to its predominant postnatal development. We also present an in vivo electroporation technique to genetically manipulate the developing cerebella and describe subsequent cerebellar analyses to assess neuronal morphology and migration.
Wprowadzenie
The cerebellum is an excellent system to study mechanisms of axon growth and migration. The cerebellum has been the subject of anatomical studies since the dawn of neuroscience1. Modern microscopy and immunohistochemical techniques have significantly expanded and refined the initial discoveries by Santiago, Ramon, and Cajal2-4. Mouse genetics and molecular studies uncovered essential growth and transcription factors in the control of cerebellar development, which led to greater understanding of crucial events required for proper wiring of different types of neurons including cerebellar granule neurons (CGNs)5-7.
The cerebellum is a derivative of rhombomere 1 of the developing hindbrain8. The rhombic lip, which is part of the roof of the 4th ventricle, gives rise to cerebellar granule neuron progenitors, which will constitute the most numerous neuronal population in the adult cerebellum9. Following rostral migration, they settle in the cerebellar anlage. Here, mitosis of granule neuron precursors leads to the dramatic expansion of the external granular layer (EGL), which takes place postnatally in rodents. From the EGL, neurons start migrating inward through the molecular layer (ML), past the Purkinje cell layer to ultimately take up residence in the internal granular layer (IGL2). During this migratory process, they acquire a bipolar shape with two axons extending into the ML. Upon further migration, the cell body migrates away from the axons and the two processes fuse to form one bifurcated, T-shaped axon10. Subsequently, these axons fasciculate and are referred to as parallel fibers. Having settled in the IGL, CGNs grow dendrites, which form dendritic claws to establish synapses with mossy fibers. To examine fundamental processes in the developing cerebellum, a combined in vitro and in vivo approach allows for reliable results and conclusions.
CGNs are not only the most numerous neurons of the cerebellum but of the entire brain and can be cultured to high purity11-13. In culture, this highly homogeneous neuronal population becomes rapidly postmitotic and acquires a polar morphology with easily identifiable axons and dendrites. Cultured CGNs have proven to be extremely useful to study various aspects of neuronal development including progenitor proliferation, differentiation, axonal and dendrite development, neuronal migration, apoptosis and electrophysiological properties (14-19 and many others). The use of genetic manipulation has expanded the versatility of cultured CGNs and allowed for further mechanistic insight into the aforementioned events. Transfection of cultured neurons using low-efficiency calcium phosphate or lipophilic methods followed by immunocytochemistry with polarity markers or software-supported analysis facilitates the assessment of e.g. the morphology of individual neurons in a dense neuronal culture. With this approach, the role of proteins of interest in axon or dendrite growth can be studied20-25,26-28. This culture system however is less useful to analyze neuronal migration as migration is very limited in high-density cultures and would require cocultures. The in vitro analysis of axon and dendrite growth also allows for the examination of interconnected proteins of a signaling pathway using combinations of RNA interference (i), over-expression or small molecules.
To establish the relevance of the protein of interest in axon and dendrite growth regulation or neuronal migration, the in vivo electroporation (IVE) technique allows for the analysis in the developing cerebellar cortex. Owing to the fact that cerebellar development in rodents extends way into the first two postnatal weeks, the cerebellum represents an accessible brain structure for genetic manipulations to examine developing axons and dendrites, neuronal migration, synaptogenesis and apoptosis20-24,29,30,26,27,31-34. In addition, this model system is also useful for other aspects of neuronal development that require the intact cerebellar cortex such as axon pathfinding, wiring and connectivity of neurons and neuron-glia interactions Taken together, this protocol provides in vitro and in vivo techniques to tackle a complementary approach regarding neuronal morphogenesis and migration.
Protokół
CGNs can be prepared either from postnatal day (P) 5 mouse pups or P6 rat pups. We follow a protocol, described by Bilimoria and colleagues, which uses a mitotic inhibitor to select for postmitotic CGNs13.
Ethics statement:
All experiments involving live animals have been conducted according to the animal protocol approved by the "Verbraucherschutz und Lebensmittelsicherheit" of Lower Saxony, Germany.
In vitro assay:
1. Preparation of DNA Plasmid, Media, and Buffers for the Calcium Phosphate Transfection Method
- Dissolve plasmid DNA in sterile, endotoxin-free water; DMEM (high glucose); make 2.5 M CaCl2; make 2x HBSS (dissolve 4 g NaCl, 0.1775 g KCl, 0.095 g Na2HPO4 •7H2O, 0.675 g glucose and 2.5 g HEPES in 250 ml ultrapure water and adjust pH to 7.05, 7.08, and 7.11). Note: When preparing the 2x HBSS solution, test which pH gives best results regarding transfection efficiency of given combination of plasmids.
2. Transfection of Cultured Neurons
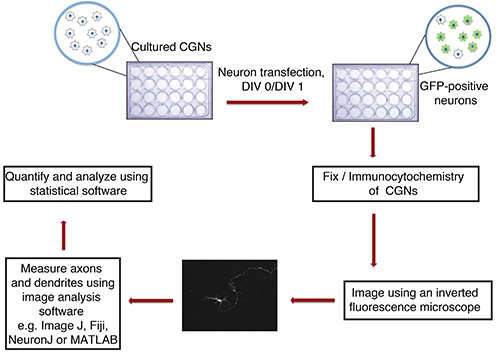
Figure 1. Flowchart of in vitro axon and dendrite growth assay. Cultured CGNs (24-well plate with glass coverslips), isolated from P6 rat pups, are transfected at DIV 0 or 1 with DNA precipitate containing a fluorescent transfection marker (e.g. GFP). After fixation and immunocytochemistry, neurons are imaged in a blinded manner. Images are imported into ImageJ and processes are measured. Measurements are then processed using a statistical program.
- Seed CGNs (20 x 106 per 24-well plate; BME, 10% calf serum, 2 mM Penicillin-Streptomycin-Glutamine (PSG), 25 mM KCl) on nitric acid-washed, polyornithine-coated 12 mm glass coverslips in a 24-well plate with 500 μl of media per well.
- On day in vitro (DIV) 0 (at least 8 hr after plating) or DIV 1, collect growth media and keep at 37 °C. Wash neurons twice with 500 μl of prewarmed DMEM and add 500 μl of DMEM.
- Place neurons in incubator (37 °C, 5% CO2) for 45 min.
- Prepare 40 μl DNA precipitate for each well by mixing: DNA (2-2.5 μg/well, 10% of total DNA should be a transfection marker e.g. GFP to visualize transfected neurons), water (up to 18 μl), add 2 μl of 2.5 M CaCl2, mix well and add 20 μl of 2x HBSS.
- Incubate DNA precipitate for 5 min at RT.
- Add DNA precipitate to each well and incubate neurons for 18 min in incubator.
- Remove DMEM/DNA mix and wash neurons twice with 500 μl of prewarmed DMEM.
- Add collected media from step 2.2 back to neurons. If neurons will be in culture for more than 3 days, supplement media with 25 mM glucose at DIV 3 to replenish carbon source.
- After 1-5 days, subject neurons to immunocytochemistry using GFP antibodies.
- Image at least 30 individual neurons per condition in a blinded manner using a fluorescent microscope.
3. Measure Axons and Dendrites with NeuronJ, an NIH ImageJ Plugin
Important: Ensure that Images are scaled correctluy by using appropriate pixel:μm ratio depending on magnification and resolution of image.
- Convert images to 8-bit with ImageJ: Open image, choose 'Image' -> 'Type' -> '8-bit' -> 'Save' image.
- Run NeuronJ plugin and open the 8-bit image.
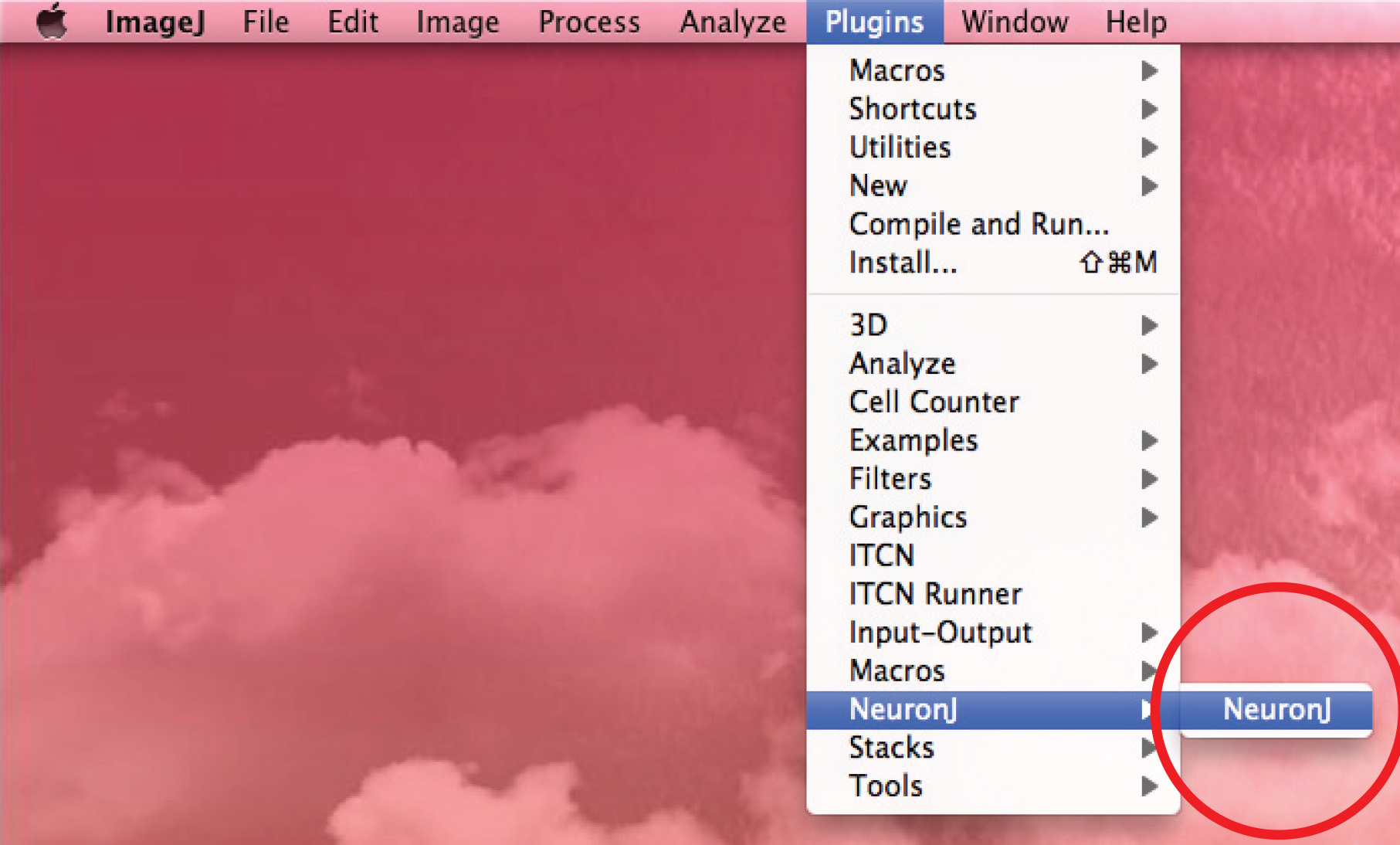
- Use the 'Add tracings' option to track the axon: click the left mouse button once at the beginning of the axon and move the mouse along the process. Double click on tip of axon if trace matches axonal shape.
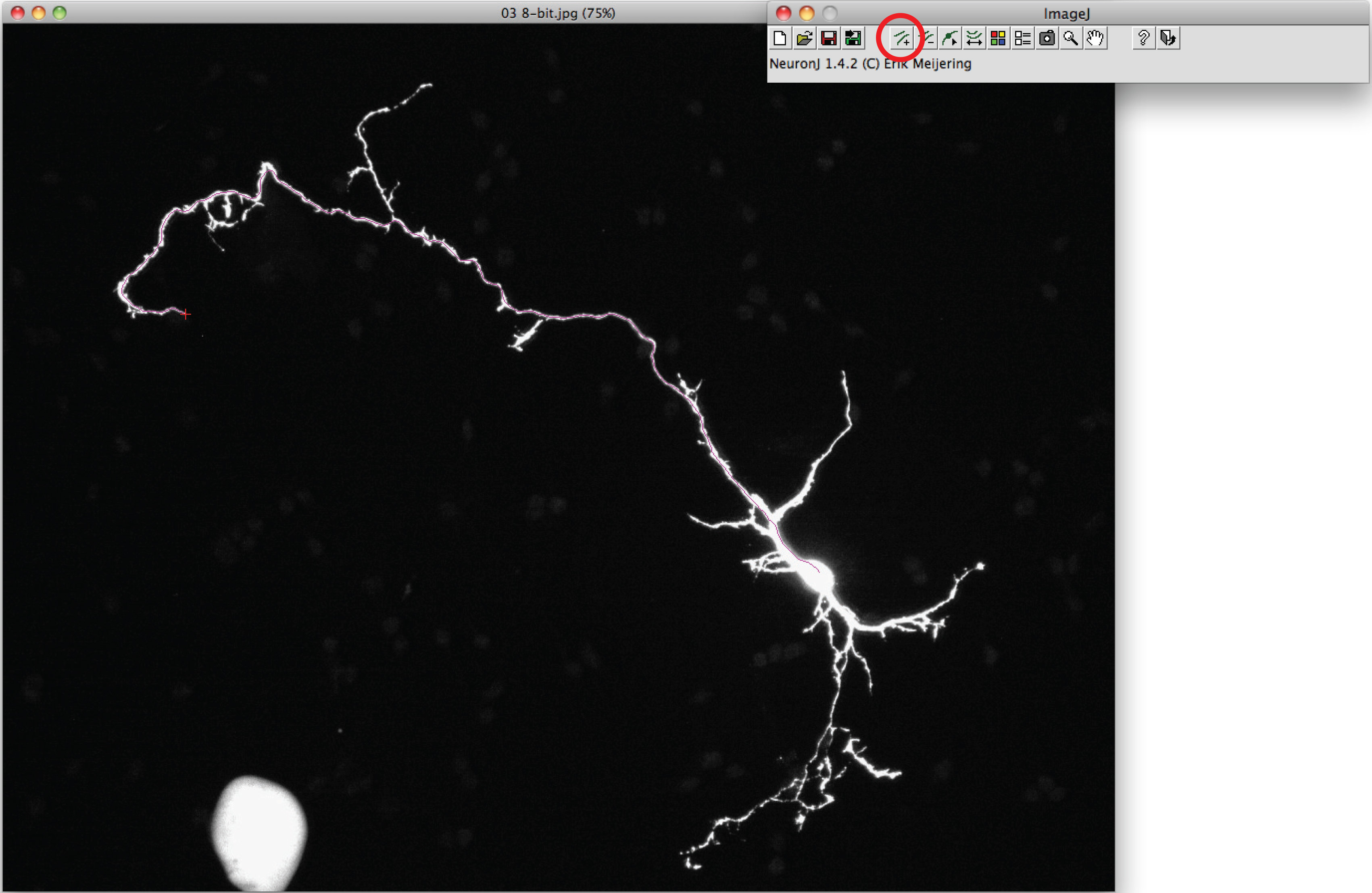
Note: Should the suggested trace differ from axonal shape, click once on the axonal process to anchor trace, then double click on tip of axon. - Click on 'Measure tracings', choose the 'Display tracing measurements' option and press 'Run'. Axon measurements are all displayed in a new window. For dendrites, choose the 'Display group measurements option' and press 'Run'.
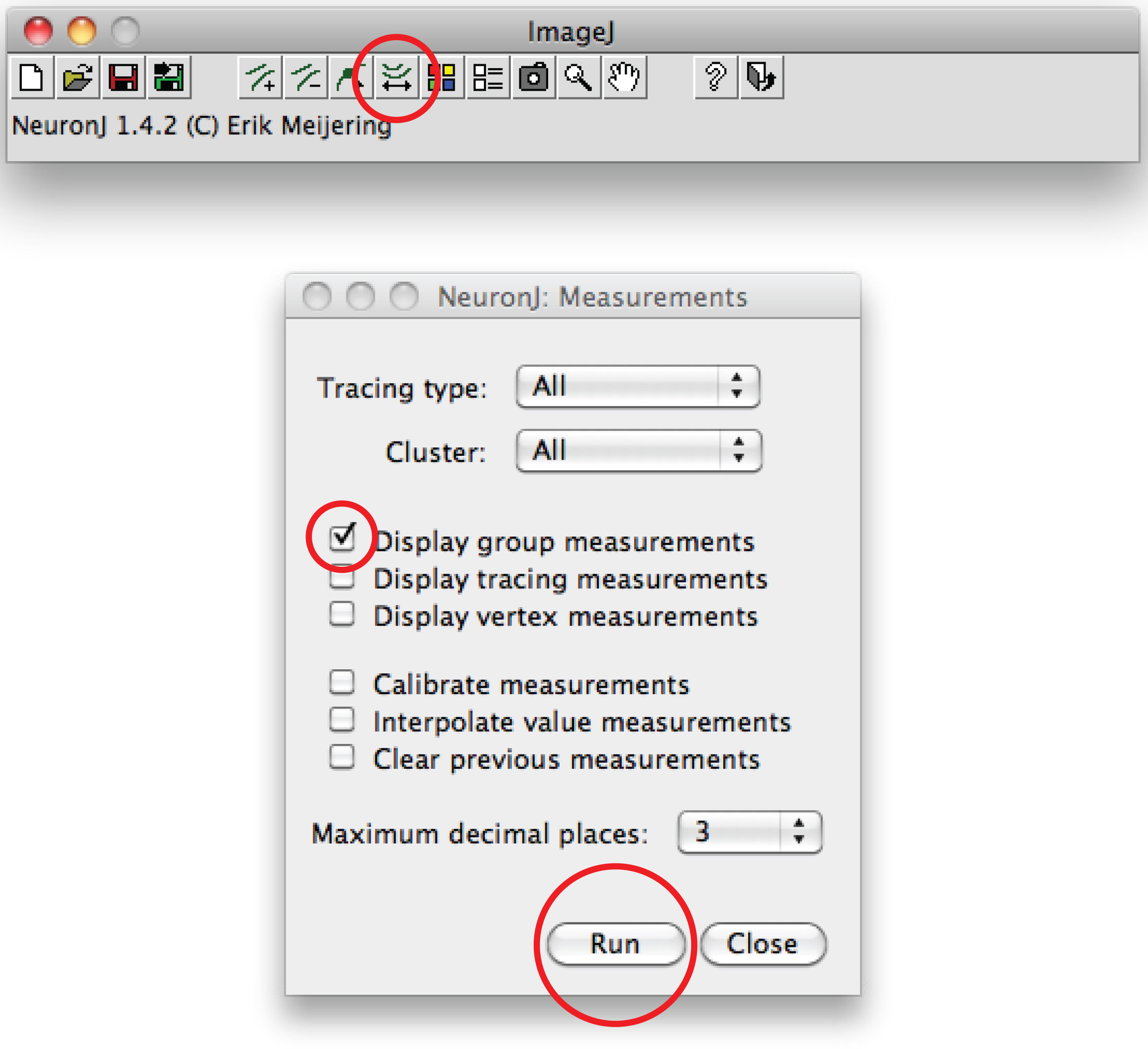
Total dendrite measurements are all displayed in a new window. Save them as a separate file that can be opened in any spreadsheet program. - Alternatively for manual tracing, use Fiji software: Right click on the 'Straight line' option, choose 'Freehand line',
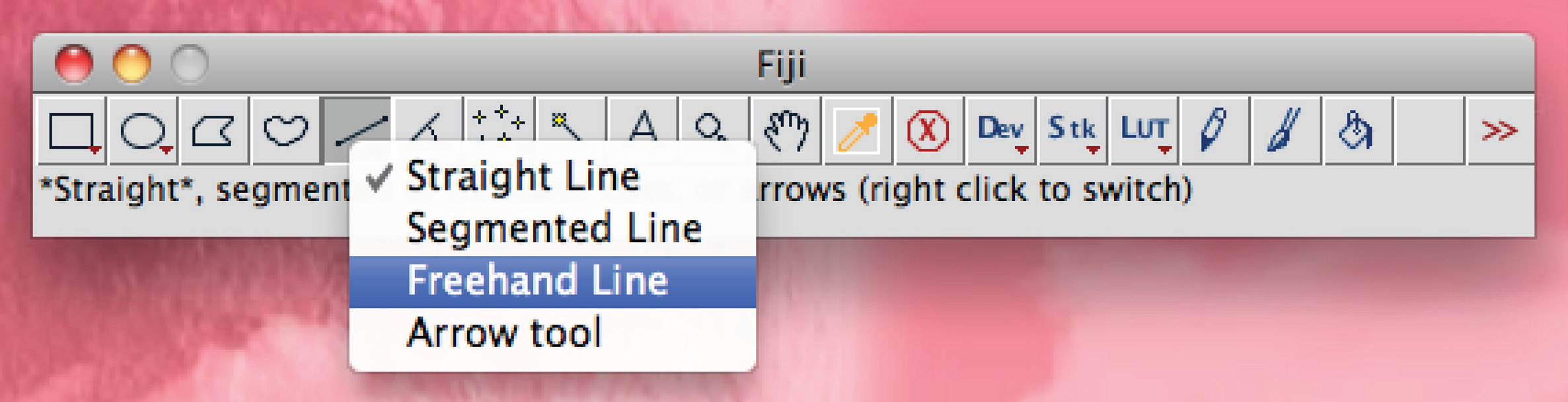
keep the left mouse button pressed and manually trace the process, press 'Ctrl+M' to measure. - Calculate average axonal/dendritic length per condition and use appropriate statistical test.
In vivo electroporation:
1. Equipment and Preparation of Reagents
- You need 30 G needles, spacer (1-2 mm), syringe, dead volume reducer (DVR), electroporator, and tweezertrodes, heating pad or infrared heat lamp, gooseneck lamp and isoflurane.
- Put DVR into needle, then attach needle to syringe, and finally put spacer onto needle (Figure 2).
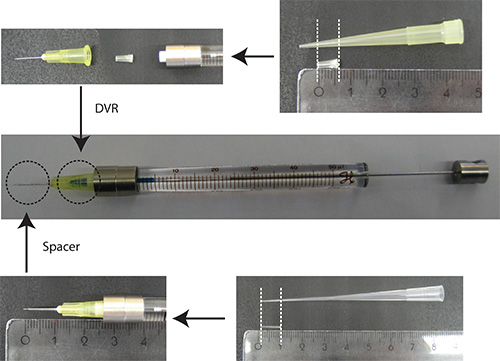
Figure 2. Preparation of needle. DVR is cut off a 200 μl pipette tip and placed into the needle to reduce the dead volume. Spacer is derived from a 200 μl loading tip and is placed on the end of the needle to regulate the depth of penetration into the cerebellum to approximately 2 mm. Ruler units: cm - Dissolve DNA in PBS/0.03% Fast Green. Note: As a transfection marker, it is advantageous to use a fluorescent protein that is under a neuron-specific promoter (e.g. Synapsin) to visualize neurons only. 25% of the total plasmid amount should be the plasmid encoding the transfection marker.
- Make 70% ethanol.
- Mix equal volumes of OCT and 30% sucrose dissolved in PBS.
- Fill syringe with 4 μl of DNA (4 μg/μl of plasmid DNA in PBS/0.03% Fast Green).
2. IVE of Rat Pups
Flowchart of IVE: see Figure 3
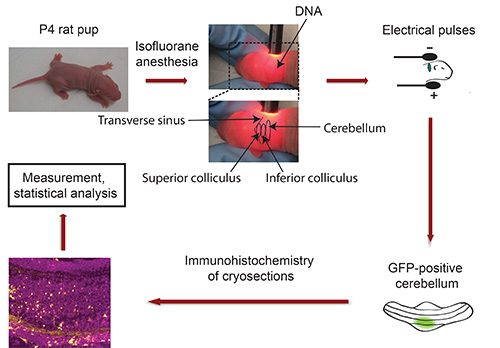
Figure 3. Flowchart of in vivo electroporation. P4 rat pups are anaesthetized with Isoflurane and plasmid DNA encoding a fluorescent transfection marker (e.g. GFP) is injected into the cerebellum, followed by exposure to 5 electrical pulses. Five days later, isolated GFP-positive cerebella are sectioned and subjected to immunohistochemistry. Images are captured using a confocal microscope and analyzed using Imaris software. Data are processed with a statistical program.
- Use P4 rat pups from albino strain (Wistar or Long Evans).
- Anesthetize pups (one after another) with isoflurane in small box (e.g. P1000 pipette tip box) with 200 μl of isoflurane (soaked in tissue) for 1-2 min until pup is no longer moving. Take care that pups do not get into contact with Iiquid isoflurane. Monitor time closely as individual pups respond differently to anesthesia.
- Sterilize back of pup’s head with 70% ethanol.
- Fix head of pup between thumb and index finger and use gooseneck lamp to locate cerebellum of albino pup. The transverse sinus sharply demarcates the midbrain (superior and inferior colliculus) from the cortical hemispheres (Figure 3). The cerebellum is located adjacent to midbrain and appears in a darker shade. Use a permanent marker to indicate the cerebellum with a dot. Important: Keep pup in fixed position! Note: Should anesthesia wear off during this procedure, expose pup to isoflurane prior to injecting the DNA.
- Insert needle (Figure 3) and slowly inject 3 μl of DNA into the cerebellum.
- Let the DNA solution diffuse for 30-60 sec.
- Place head of pup between tweezertrodes so that the minus pole makes contact with the back of the head (cerebellar region) and that the plus pole contacts the opposite side of the head (Figure 3).
- Subject pup to 5 electrical pulses. Adjust voltage to weight of pups to ensure good electroporation efficiency without compromising their survival (Table 1).
Table 1. Electroporation of P4 rat pups.Weight Voltage Pulse Interval 8-9 g 160 V 50 msec 950 msec 9-10 g 165 V 50 msec 950 msec > 10 g 170 V 50 msec 950 msec - Let pups recover on heated pad or underneath an infrared lamp. Return pups to dam. Important: Make sure that heating source does not inflict any burns.
- Sacrifice pups 5 days after electroporation by placing them in CO2 followed by decapitation.
- Isolate the cerebella and screen for GFP-positive ones cerebella using a fluorescent microscope.
- Fix cerebella in 4% PFA O/N at 4 °C, then incubate in 30% sucrose at 4 °C until cerebella sink to bottom of tube.
- Embed cerebella in OCT/30% sucrose and cut 40 μm coronal sections using a cryostat. Note: Section each cerebellum in a blinded manner.
- Subject sections to immunohistochemistry using the GFP antibody. Counterstain with nuclear dye (DAPI or Hoechst 33258) and determine localization of at least 200 transfected neurons per animal.
- For an in-depth analysis, subdivide the IGL into halves, resulting in an upper IGL facing the ML and a lower IGL facing the white matter and count GFP-positive neurons residing in each half. Note: Count GFP-positive neurons of each section in a blinded manner.
3. Measuring Dendrite Length, Acquire the Images of the Section in x, y, z Plane Using a Confocal Microscope
Note: for example, use 40 images for a 40 μm section with a z-stwp of 1 μm.
- Open image series in the software, Imaris, to generate a 3D image of the dendrites.
- Click on 'Surpass' mode to view the neuron in 3D.
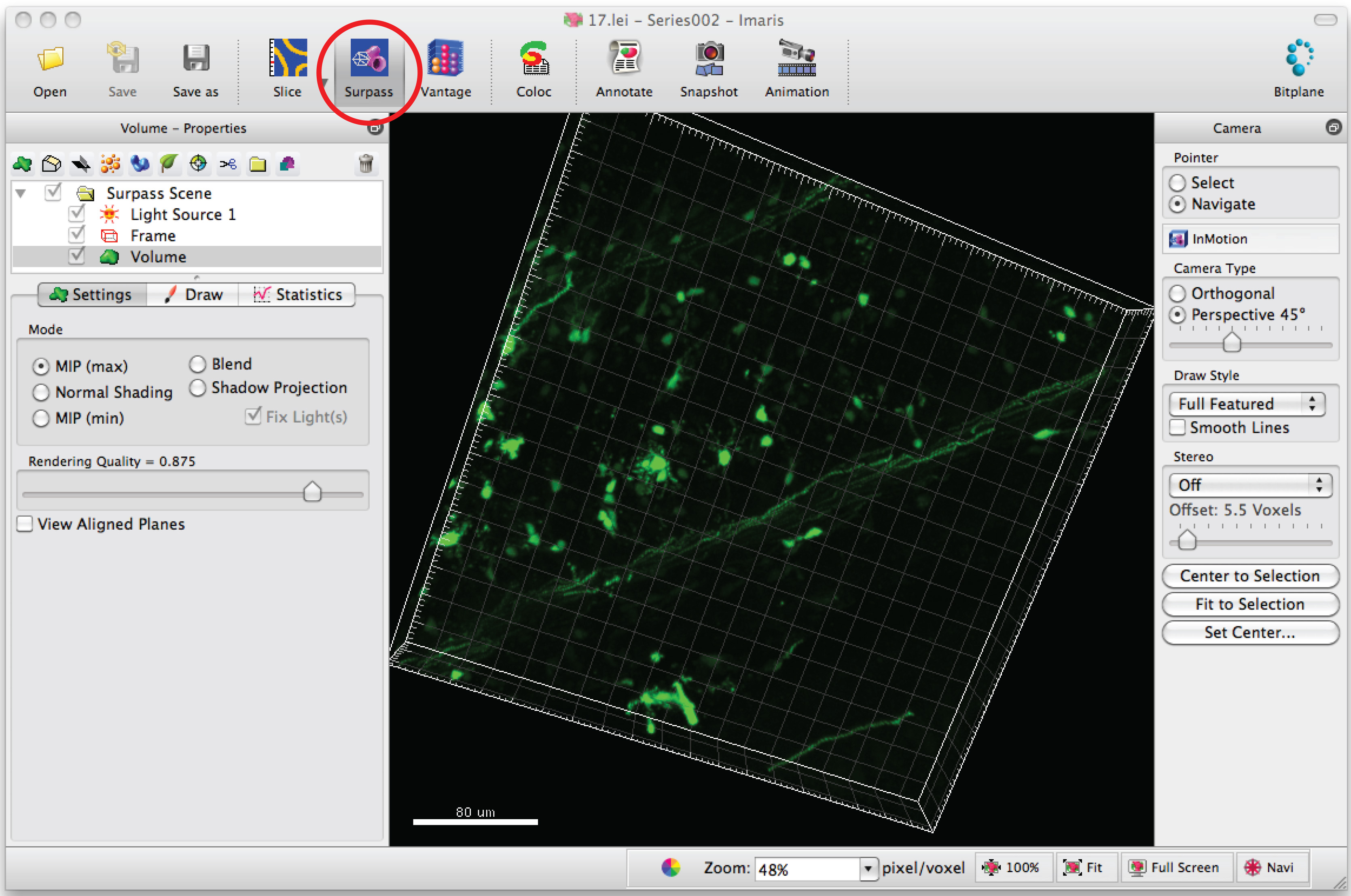
- Select 'Add new filament' and click 'Skip automatic creation' to start semiautomatic tracing.

Note: Analyze each 3D image in a blinded manner - Select 'Draw' tab and 'AutoPath'.
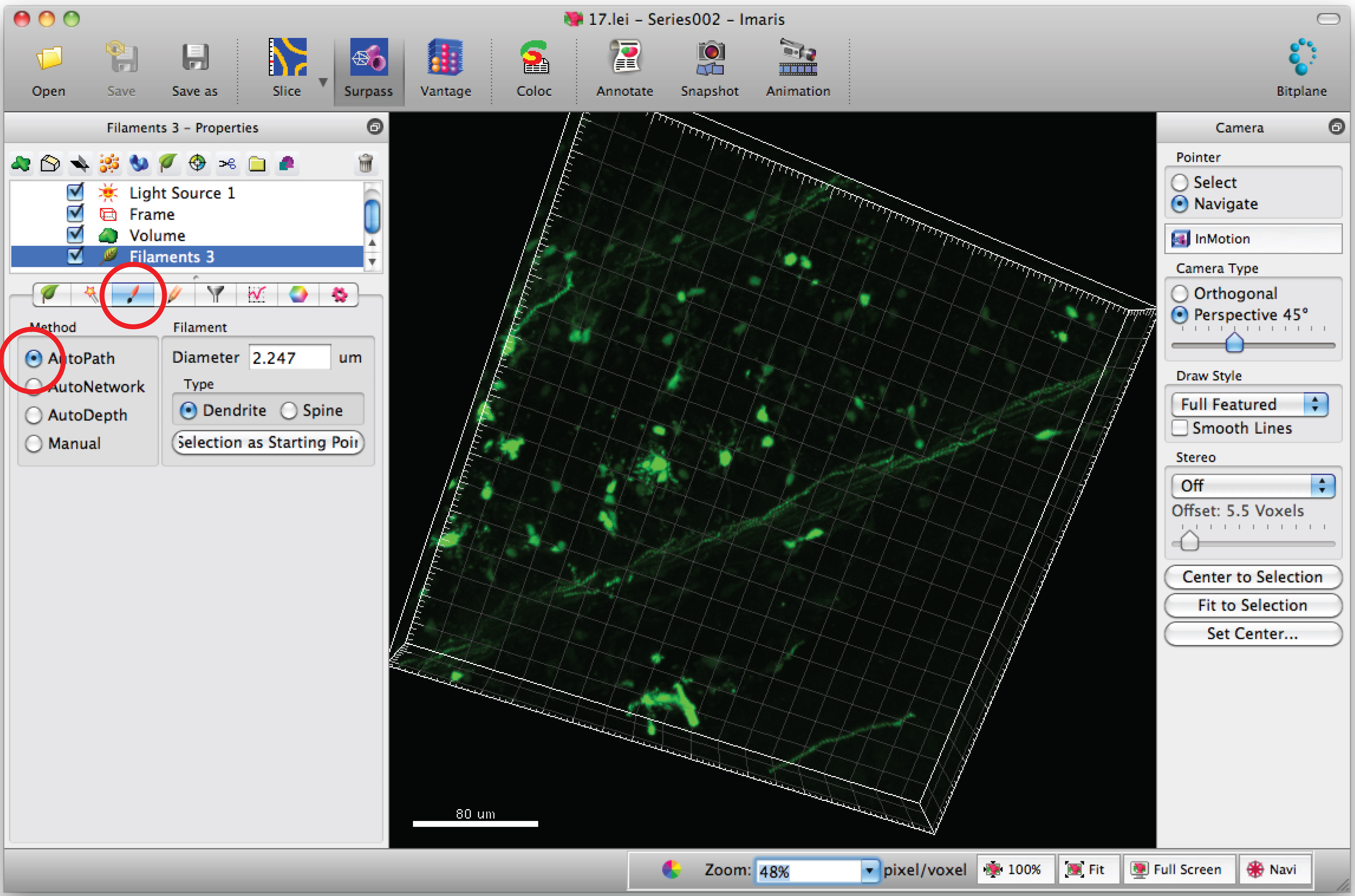
- Move the mouse cursor on the cell body and Shift+mouse right click to select the cell body. Note: Autocalculation by software may require a few minutes.

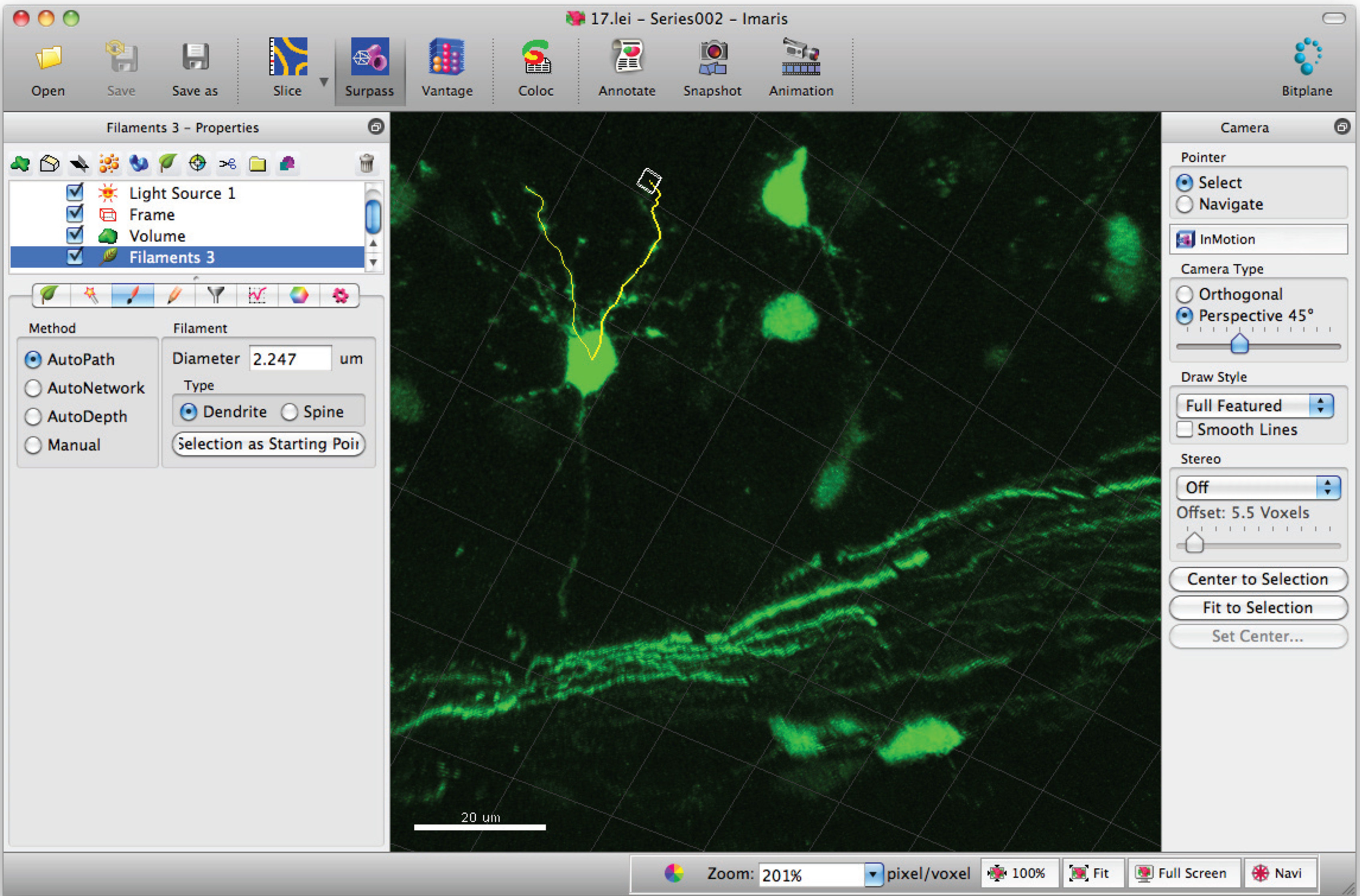
- Add paths to the filament (dendrite) using Shift+mouse left click. Note: Paths can be visualized in real time.
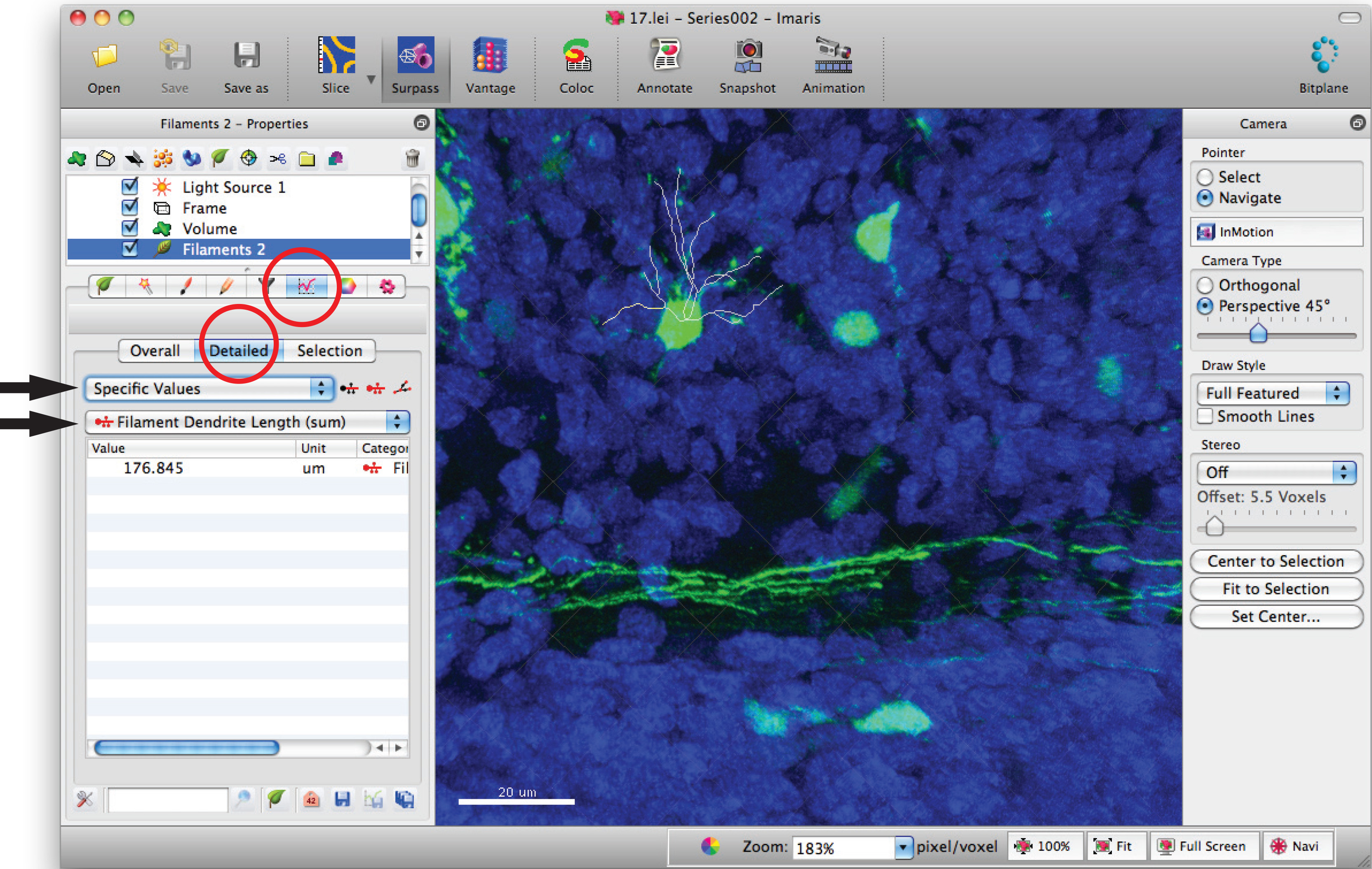
- Go to filament statistics window and click on 'Detailed', 'Specific values' and 'Filament dendrite length (sum)' for sum of total dendrite length.
- Use appropriate statistical tests to analyze data.
Wyniki
To analyze the morphology of CGNs in response to different culturing conditions, we transfected the neurons on DIV 0 as described above. After transfection, we placed one set of neurons into full medium (BME, 10% calf serum, 2 mM PSG, 25 mM KCl) and another set into minimal medium containing insulin (BME, 25 mM glucose, 2 mM PSG, 10 μg/ml insulin). We subjected the neurons to immunocytochemistry using the GFP antibody at DIV 1, 2, and 3, followed by measuring axons and dendrites for set 1 and axons only for set 2. O...
Dyskusje
Advantages and limitations of the described in vitro and in vivo methods:
Cultured CGNs from mouse and rats are equally well suited for morphological analyses. Owing to the bigger size of a rat cerebellum, the yield of CNGs from rat pups exceeds that of mouse pups 3-4x. Aside from CGNs, cortical and hippocampal neurons can be used as culture system as well. The calcium phosphate method results in a low (0.01-5%) transfection efficiency, which is desired to analyze the morphol...
Ujawnienia
The authors declare no competing financial interests.
Podziękowania
We thank N. Schwedhelm-Domeyer for excellent technical assistance, C. Hammer and S. Papiol for help with statistical analyses. Our work is funded by the Max Planck Society, the Deutsche Forschungsgemeinschaft, the Center for Nanoscale Microscopy, and Molecular Physiology of the Brain (CNMPB), Göttingen, Germany and by the GGNB Junior Group Stipend of the University of Göttingen.
Materiały
| Name | Company | Catalog Number | Comments |
| DMEM | Gibco | 11960-044 | |
| BME | Gibco | 41010-026 | |
| Insulin | Sigma-Aldrich | Si-1-4011 | |
| Poly-L-Ornithine | Sigma-Aldrich | P-2533 | |
| CaCl2 | Appli-Chem | A3652 | |
| Isoflurane | Actavis Deutschland | ||
| Tissue-Tek OCT | Sakura | ||
| ECM 830 and tweezertrodes | Harvard Apparatus | ||
| Epifluorescence microscope and camera | Nikon | ||
| SP2 confocal microscope | Leica | ||
| ImageJ | NIH | ||
| Imaris 7.4.2 | Bitplane, Inc. | ||
| GraphPad Prism | GraphPad Software, Inc. | ||
| MS Excel | Microsoft | ||
| Loading tip 1-200 µl | Costar | 4853 | |
| Pipette tip 200 µl | Sarstedt | 70.760.502 | |
| Microlance 3 needle, 30 G | BD | 302200 | |
| 50 µl gastight Syringe 1705 | Hamilton | ||
| Glass coverslips | Thermo Scientific Menzel Glaeser | CB00120RA1 |
Odniesienia
- Cajal, S. R. . Histology of the nervous system of man and vertebrates. , (1995).
- Altman, J., Bayer, S. A. . Development of the cerebellar system : in relation to its evolution, structure, and functions. , (1997).
- White, J. J., Reeber, S. L., Hawkes, R., Sillitoe, R. V. Wholemount immunohistochemistry for revealing complex brain topography. J. Vis. Exp. , (2012).
- Palay, S. L., Chan-Palay, V. . Cerebellar cortex: cytology and organization. , (1974).
- Hatten, M. E., Alder, J., Zimmerman, K., Heintz, N. Genes involved in cerebellar cell specification and differentiation. Curr. Opin. Neurobiol. 7, 40-47 (1997).
- Hatten, M. E., Heintz, N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu. Rev. Neurosci. 18, 385-408 (1995).
- Sillitoe, R. V., Joyner, A. L. Morphology molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Ann. Rev. Dev. Biol. 23, 549-577 (2007).
- Zervas, M., Millet, S., Ahn, S., Joyner, A. L. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 43, 345-357 (2004).
- Wingate, R. J. The rhombic lip and early cerebellar development. Curr. Opin. Neurobiol. 11, 82-88 (2001).
- Kawaji, K., Umeshima, H., Eiraku, M., Hirano, T., Kengaku, M. Dual phases of migration of cerebellar granule cells guided by axonal and dendritic leading processes. Mol. Cell Neurosci. 25, 228-240 (2004).
- Hatten, M. E., Gao, W. -. Q., Morrison, M. E., Mason, C. A. Cellular and molecular neuroscience. , 419-41 .
- Lee, H. Y., Greene, L. A., Mason, C. A., Manzini, M. C. Isolation and culture of post-natal mouse cerebellar granule neuron progenitor cells and neurons. J. Vis. Exp. , (2009).
- Bilimoria, P. M., Bonni, A. Cultures of cerebellar granule neurons. CSH Protoc. 2008, (2008).
- Rivas, R. J., Hatten, M. E. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J. Neurosci. 15, 981-989 (1995).
- Kuhar, S. G., et al. Changing patterns of gene expression define four stages of cerebellar granule neuron differentiation. Development. 117, 97-104 (1993).
- Baird, D. H., Hatten, M. E., Mason, C. A. Cerebellar target neurons provide a stop signal for afferent neurite extension in vitro. J. Neurosci. 12, 619-634 (1992).
- Segal, R. A., Pomeroy, S. L., Stiles, C. D. Axonal growth and fasciculation linked to differential expression of BDNF and NT3 receptors in developing cerebellar granule cells. J. Neurosci. 15, 4970-4981 (1995).
- Gallo, V., Ciotti, M. T., Coletti, A., Aloisi, F., Levi, G. Selective release of glutamate from cerebellar granule cells differentiating in culture. Proc. Natl. Acad. Sci. U.S.A. 79, 7919-7923 (1982).
- Smith, T. C., Wang, L. Y., Howe, J. R. Distinct kainate receptor phenotypes in immature and mature mouse cerebellar granule cells. Physiol. 517 (1), 51-58 (1999).
- Konishi, Y., Stegmuller, J., Matsuda, T., Bonni, S., Bonni, A. Cdh1-APC controls axonal growth and patterning in the mammalian brain). Science. 303, 1026-1030 (2004).
- Stegmuller, J., Huynh, M. A., Yuan, Z., Konishi, Y., Bonni, A. TGFbeta-Smad2 signaling regulates the Cdh1-APC/SnoN pathway of axonal morphogenesis. J. Neurosci. 28, 1961-1969 (2008).
- Stegmuller, J., et al. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 50, 389-400 (2006).
- Kannan, M., Lee, S. J., Schwedhelm-Domeyer, N., Nakazawa, T., Stegmuller, J. p250GAP is a novel player in the Cdh1-APC/Smurf1 pathway of axon growth regulation. PLoS One. 7, (2012).
- Kannan, M., Lee, S. J., Schwedhelm-Domeyer, N., Stegmuller, J. The E3 ligase Cdh1-anaphase promoting complex operates upstream of the E3 ligase Smurf1 in the control of axon growth. Development. 139, 3600-3612 (2012).
- Gaudilliere, B., Konishi, Y., de la Iglesia, N., Yao, G., Bonni, A. A. CaMKII-NeuroD Signaling Pathway Specifies Dendritic Morphogenesis. Neuron. 41, 229-241 (2004).
- Yang, Y., et al. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 326, 575-578 (2009).
- Litterman, N., et al. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 9, (2011).
- Kim, A. H., et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 136, 322-336 (2009).
- Shalizi, A., et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 311, 1012-1017 (2006).
- Vadhvani, M., Schwedhelm-Domeyer, N., Mukherjee, C., Stegmuller, J. The Centrosomal E3 Ubiquitin Ligase FBXO31-SCF Regulates Neuronal Morphogenesis and Migration. PLoS One. 8, (2013).
- Puram, S. V., et al. A CaMKIIbeta signaling pathway at the centrosome regulates dendrite patterning in the brain. Nat. Neurosci. , (2011).
- Jia, Y., Zhou, J., Tai, Y., Wang, Y. TRPC channels promote cerebellar granule neuron survival. Nat. Neurosci. 10, 559-567 (2007).
- Huynh, M. A., et al. An isoform-specific SnoN1-FOXO1 repressor complex controls neuronal morphogenesis and positioning in the mammalian brain. Neuron. 69, 930-944 (2011).
- de la Torre-Ubieta, L., Bonni, A. Transcriptional regulation of neuronal polarity and morphogenesis in the mammalian brain. Neuron. 72, 22-40 (2011).
- Calissano, P., et al. Recombinant human insulin-like growth factor I exerts a trophic action and confers glutamate sensitivity on glutamate-resistant cerebellar granule cells. Proc. Natl. Acad. Sci. U.S.A. 90, 8752-8756 (1993).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone