Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Simple Method for Fluorescence DNA In Situ Hybridization to Squashed Chromosomes
W tym Artykule
Podsumowanie
Here, we present a simple method for performing fluorescence DNA in situ hybridization (DNA ISH) to visualize repetitive heterochromatic sequences on slide-mounted chromosomes. The method requires minimal reagents and it is versatile for use with short or long probes, different tissues, and detection with fluorescence or non-fluorescence-based signals.
Streszczenie
DNA in situ hybridization (DNA ISH) is a commonly used method for mapping sequences to specific chromosome regions. This approach is particularly effective at mapping highly repetitive sequences to heterochromatic regions, where computational approaches face prohibitive challenges. Here we describe a streamlined protocol for DNA ISH that circumvents formamide washes that are standard steps in other DNA ISH protocols. Our protocol is optimized for hybridization with short single strand DNA probes that carry fluorescent dyes, which effectively mark repetitive DNA sequences within heterochromatic chromosomal regions across a number of different insect tissue types. However, applications may be extended to use with larger probes and visualization of single copy (non-repetitive) DNA sequences. We demonstrate this method by mapping several different repetitive sequences to squashed chromosomes from Drosophila melanogaster neural cells and Nasonia vitripennis spermatocytes. We show hybridization patterns for both small, commercially synthesized probes and for a larger probe for comparison. This procedure uses simple laboratory supplies and reagents, and is ideal for investigators who have little experience with performing DNA ISH.
Wprowadzenie
DNA in situ hybridization (DNA ISH) is a commonly used method for mapping sequences to specific chromosome regions. Probes to single copy regions within euchromatin can be generated through a handful of approaches, including nick-translation or end-labeling of long DNA products1,2 and the incorporation of deoxygenin (DIG)-attached nucleotides and their recognition through a wide variety of group-conjugated antibodies1-3. Visualization of euchromatic sequences in few or single copy number requires the use of either single, large probes with high specific activity or a cocktail of multiple, smaller probes that collectively enhance signal.
In contrast, highly repetitive sequences found in heterochromatin, such as satellite DNAs, are easier targets for DNA ISH because they normally exist as tens to thousands of repeats clustered in single chromosome regions known as blocks. Transposable elements also can be found at high copy numbers at distinct chromosomal loci2. In these cases, single probes with low specific activity can effectively label heterochromatic sequences due to their hybridization at multiple sites. Probes to repetitive sequences can be synthesized commercially as short oligonucleotides (30-50 bp) and chemically conjugated with any of multiple different fluorescent groups. Mapping repetitive sequences within heterochromatin by using genome-sequencing technologies is difficult due to challenges encountered in building scaffolds within highly repetitive satellite blocks4-6,7. Currently, ISH stands as the most effective way of mapping these sequences at the sub-chromosome level. This strategy is important for mapping large numbers of repetitive sequences that are being uncovered by ongoing genome and transcriptome sequencing studies.
The efficiency and ease of mapping repetitive sequences on slide-mounted chromosomes would be greatly enhanced by a simplified protocol for DNA ISH. For example, existing protocols for DNA ISH involve multiple washes of hybridized tissues in formamide solution2,8, thus adding substantially to the required time for mapping sequences and also producing large amounts of chemical waste for this costly reagent. Here we describe a revised DNA ISH method that circumvents the need for formamide washes and utilizes basic laboratory equipment and reagents. This method was originally designed for the rapid mapping of highly repetitive DNA sequences in heterochromatic regions of Drosophila larval neuroblasts by using commercially synthesized oligos that are conjugated with fluorescence dyes. However, this method also works for mapping repetitive sequences by using larger probes synthesized through other means9,10 and across multiple different tissue and chromosome types. Additionally, this method can be used to map euchromatic sequences by using longer or multiple, short probes within the euchromatic sequence of interest.
Protokół
1. Tissue Dissection and Fixation (60 min)
- For Drosophila brains, place 3rd instar larvae in a drop of 1x PBS (phosphate buffered saline). Choose large 3rd instar larvae that are actively crawling from vials or bottles that are not overcrowded.
- Use one ultrafine tweezer pair to grab hold of the mouth hooks and another tweezer pair to grab 2/3 down the length of the body (Figure 1A, B). Pull gently on the mouth hooks to expose the brain, ventral ganglia, salivary glands and part of the larval digestive tract. Use the tweezers to separate the brain and ventral ganglia (Figure 1A, B) from the other tissues and place in a droplet of 1x PBT (phosphate buffered saline with Tween) on a plastic Petri plate.
- For Nasonia testis dissections, choose male 3-day-old pupae (yellow bodies with red eyes). Male Nasonia have small wing pad lengths relative to females during the pupal stage (Figure 1C).
- Hold the pupa at the top of the abdomen near the thoracic region with one tweezer pair, and using the other tweezer pair, grab the very distal tip of the abdomen and pull out the tear-drop-shaped testes (they will be surrounded by fat body that can be gently shaken away; Figure 1D). Detach any exterior body parts from the testes, which will prevent proper squashing, and place in a droplet of 1x PBT on a plastic Petri plate.
- For each slide, dissect a total of four or five tissue samples (i.e., Drosophila larval brains or Nasonia testes).
NOTE: More than five samples will lead to overcrowded nuclei and chromosomes. - Optionally, to achieve some separation of sister chromatids in euchromatic regions of mitotic chromosomes, treat the tissue with a hypotonic solution: transfer brains from 1x PBT to a drop of 0.5% sodium citrate for 5-10 (no more than 10) min.
NOTE: Colchicine (a mitotic inhibitor) can be helpful for increasing the number of mitotic figures at metaphase11. However, its use is unnecessary if very large healthy larvae are used, and even undesirable because it can negatively affect the chromosome resolution, spreading and morphology. - Place a droplet (~20 µl) of fixative solution (2.5% paraformaldehyde in 45% acetic acid) onto the surface of a clean Sigmacote-treated cover slip.
NOTE: Make fixative solution fresh for each day of use. Fixative solutions ranging from 1.8-3.7% paraformaldehyde in 45% acetic acid yield the best results for FISH. Using tissues other than brains or testes, or adapting this protocol for immuno-FISH may require experimenting with different fixatives (for a list of different fixatives, see12). - Carefully transfer each tissue sample from the dissecting buffer (1x PBT) into the fixative droplet with ultrafine tweezers, minimizing the transfer of dissecting buffer into the fixative solution. Position the tissue samples so that they are evenly spaced from one another within the fixative droplet. Incubate the tissues in fixative for 4 min at room temperature.
- Carefully place a poly-lysine-coated slide face down onto the tissue and cover slip. Do not press down at this point but instead allow the two to contact lightly so that the cover slip sticks onto the underside of the slide. Invert the slide so that the cover slip is on top.
- Sandwich the slide/tissue/cover slip inside a folded piece of filter paper. On a stable surface, using the thumb, press very firmly straight down onto the position directly above the cover slip. Be careful to avoid lateral sliding of the cover slip (this will cause smearing of the tissue).
- Submerge the slide/tissue/cover slip into liquid nitrogen (see apparatus in Figure 1E, F), and let stand until the Nitrogen stops boiling (longer is fine). Remove the slide and immediately snap off the cover slip with a fresh razor blade by flicking a corner of the cover slip in an upward direction (avoid scratching the fixed tissue with the razor blade).
- Pre-cooling the slide/tissue/cover slip on a block of dry ice, cover slip up, for 1-2 min prior to submerging in liquid nitrogen will help prevent the slides from cracking.
- Immediately place the slide with tissue into a Coplin jar filled with 100% ethanol at room temperature and let stand for at least 5 min (this time can be longer if needed).
NOTE: Cold 100% ethanol could also be used. - Remove the slide with tissue, wick away excess ethanol with a Kimwipe (without touching the fixed tissue), and leave the slide to air dry for 1 hr.
- Proceed directly to step 2 or keep slides dry in low humidity air or in a desiccation chamber for weeks to even months before performing the hybridization.
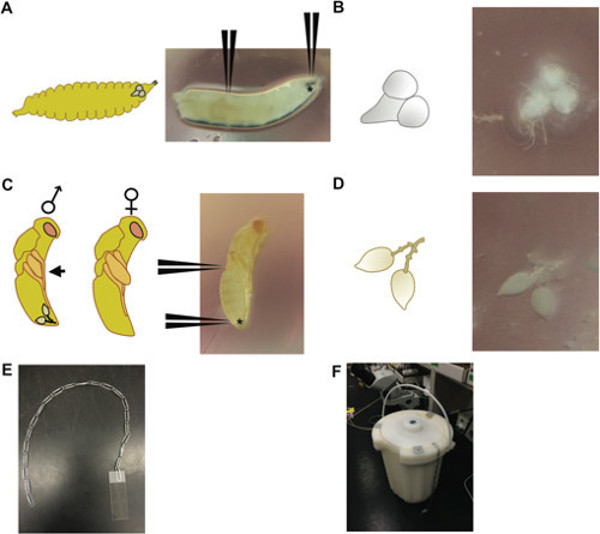
Figure 1: (A) A 3rd instar Drosophila larva (right), with positions indicated for where to grab the mouth hooks (denoted with *) and 2/3 of the way down the larvae to dissect brains; (left) a schematic of a larva a the same developmental stage, depicting the relative position of the brain within the larval head. (B) The brain and ventral ganglia dissected from a 3rd instar Drosophila larva (right) and a schematic of this tissue (left). (C) 3-day-old Nasonia pupa at the yellow body-red eye stage. (D) A testis pair dissected from a 3 day old Nasonia male pupa (right) with positions indicating where to grab the pupa at the posterior of the abdomen (denoted with *) and midway on the body; (left) schematic depicting male and female wasp pupae; male pupae can be distinguished by wings that do not extend past the saggital profile (black arrow), in contrast to females, which have wings that extend past the profile; the relative position of the testis pair is shown in the male pupa. (E) The apparatus—a string of paperclips—and (F) method used to immerse slides in a vessel of liquid nitrogen. Multiple paperclip strings can be used for simultaneous immersion of multiple slides.
2. In situ Hybridization (30 min on day 1; 1 hr—2.5 hr for long probes—on day 2)
- Add 1 µl (100 ng) of each probe to 20 µl of 1.1x hybridization buffer. Pipet probe/hybridization buffer onto the surface of the fixed tissue (avoid touching the tissue).
- Carefully place a cover slip directly onto the probe/hybridization buffer, making sure that the cover slip is centered directly over the tissue. The buffer should migrate to the outer edge of the cover slip, leaving no air bubbles.
NOTE: Small bubbles that do not contact tissue do not pose any problems to the procedure. Remove large bubbles by carefully lifting a corner of the cover slip and carefully dropping it back onto the slide. - Place the slide/tissue/cover slip onto the surface of a block pre-heated to 95 °C (cover slip up). Cover with a large piece of aluminum foil to prevent light exposure. Let the slide incubate at 95 °C for 5 min.
NOTE: A typical heat block with holes for tubes can be flipped over to provide a flat surface on which to place the slide/tissue/cover slip. - Remove the slide, letting it cool slightly until it is warm to the touch. Carefully wrap a piece of stretched Parafilm around the cover slip to seal the liquid beneath it.
- Place the sealed slide inside a humidity chamber and place the chamber into an incubator preheated to 30 °C. Incubate at 30 °C for 4 hr to overnight.
- Create a humidity chamber from an empty tip box or lidded Tupperware container with dampened Kimwipes or paper towels placed at the bottom.
NOTE: Single stranded DNA oligo probes have been designed to be 28-33 bases to achieve a theoretical melting temperature (Tm) of 45-47 °C. These length and Tm ranges reflect the fact that many repetitive sequences that we have studied are AT-rich and therefore have very low GC content. Longer probes will likely have higher Tm values; this may result in higher background hybridization at the standard hybridization temperature of 30 °C. Thus, some troubleshooting with hybridization temperatures may be required to achieve the best results. To find the best hybridization temperature, increase (or decrease) the temperature by 5 °C, incrementally.
- Create a humidity chamber from an empty tip box or lidded Tupperware container with dampened Kimwipes or paper towels placed at the bottom.
- Carefully remove the Parafilm from the slide and then carefully remove the cover slip by slowly lifting one corner. Wash the slide three times for 15 min each wash in 0.1x SSC buffer. Cover the Coplin jar with aluminum foil during the washes to minimize light exposure.
- If not using a long biotinylated probe, proceed to step 2.8.
- If using a long biotinylated probe, dry the area around the tissue with a Kimwipe, being careful not to touch the tissue itself. Place 100 µl of blocking solution over the tissue and cover gently with a coverslip, taking care to avoid trapping bubbles. Wrap the slide over the coverslip with Parafilm and place at 37 °C for 30 min.
- Carefully remove the coverslip and blot around the tissue with a Kimwipe. Pipet 100 µl of rhodamine-avidin diluted 1:1,000 in SBT over the tissue and cover gently with a coverslip, taking care to avoid trapping bubbles. Wrap the slide over the coverslip with Parafilm and place at 37 °C for 30 min.
- Carefully remove the coverslip and wash the slide 3 times for 5 min each in 4x SSCT and then 3 times for 5 min each in 0.1x SSC.
NOTE: Slides can be washed for longer periods of time.
- Remove the slide and blot around the tissue with a dry Kimwipe to remove excess buffer (avoid touching the tissue). Place the slide tissue side up in a dark place for 10-15 min or until the moisture completely dissipates.
- Pipet 11 µl of Vectashield mounting medium (with 4’,6-diamidino-2-phenylindole—DAPI) onto the tissue. Carefully place a clean cover slip (not treated with Sigmacote) directly over the center of the mounting medium and tissue. The mounting medium should migrate slowly outward toward the edges of the cover slip.
NOTE: If the mounting medium fails to reach the edge of the cover slip on all sides, then an additional 1-2 µl of mounting medium can be applied to one position at the edge of the cover slip to fill in the needed volume. In this case, be sure to wipe away any excess medium from the slide surface before sealing. - Seal the edges of the cover slip with nail polish. Avoid painting the nail polish over the tissue sample.
- Place the slide upright in a dark place and let the nail polish dry until completely hard (usually 30 min or more). At this point, image the tissue or store at -20 °C for up to 1 week for later imaging.
Buffer/Solution Recipes
10x PBS
- 80 g NaCl
- 2.0 g KCl
- 14.4 g Na2HPO4
- 2.4 g KH2PO4
- pH to 7.4, H2O to 1 L
1x PBT
- 5 ml 10x PBS
- 45 ml H2O
- 0.1% Tween 20
20x SSC
- 175.3 g NaCl
- 88.2 g Na Citrate
- in 800 ml H2O
- pH to 7, H2O to 1 L
4x SSCT
- 200 ml 20x SSC
- 799 ml H2O
- 0.1% Tween 20
0.1x SSC
- 5 ml 20x SSC
- 995 ml H2O
Hybridization mix (20 µl; modified from 11)
- 10 µl formamide
- 4 µl 50% dextran sulfate
- 2 µl 20x SSC
- 4 µl H2O
SBT8 (10 ml)
- 2 ml 20x SSC
- 0.01 g Bovine Serum Albumin (BSA)
- 10 µl Tween 20
- 7.9 ml H2O
Blocking solution8 (10 ml)
- 0.3 g BSA
- 10 µl Tween 20
- 2 ml 20x SSC
- 8 ml H2O
Fixative solution with paraformaldehyde (1 ml)
- 393.75 µl H2O (add water first)
- 450 µl glacial acetic acid
- 156.25 µl 16% paraformaldehyde
Wyniki
To demonstrate this method, we hybridized a set of small commercially synthesized oligos that were chemically modified with fluorescent conjugates (Figure 2) and a longer biotinylated probe (made through nick translation of a PCR product; Figure 2B), to chromosomes from several different tissue types (see Table 1). The target sequences included satellite repeats located in pericentromeric (heterochromatic) regions of mitotic chromosomes from D. melanogaster larv...
Dyskusje
DNA ISH is frequently used to map specific sequences to chromosomes. We have described a simple method for DNA ISH optimized for high copy number, heterochromatic sequences. Rather than using washes in a formamide solution, which is a requirement in other existing DNA ISH protocols, we place tissue-mounted slides directly on a pre-heated block to denature DNA. This method circumvents the use of large amounts of formamide. One critical step for producing crisp hybridization signals is to use freshly made fixation solution...
Ujawnienia
The authors declare that they have no competing financial or any other conflict of interest.
Podziękowania
We thank Zhaohua Irene Tang in the W. M. Keck Science Department for the use of her epifluorescence microscope and the Werren lab for donating Nasonia for dissections. This work was supported in part by an NIH-NRSA fellowship (5F32GM105317-02) to AML.
Materiały
| Name | Company | Catalog Number | Comments |
| Poly-L-lysine coated slides (regular slides also can be used) | Sigma Aldrich | ||
| Ultrafine tweezers (5 gauge) | Dumont | ||
| 22 x 22 mm cover slips | Fisher | Sigmacote-treated by immersion for 15 sec, blotting dry, and wiping away all traces of Sigmacote so that cover slip is clear | |
| Sigmacote | Sigma | ||
| Filter paper | 75 - 150 mm | ||
| Paraffin wax paper | |||
| Heat block with thermometer | |||
| Dry incubator | |||
| Razor blades | |||
| Humidity chamber | empty pipette tip box or Tupperware, lined with moistened paper towels or Kimwipes | ||
| Coplin jars | with slide grooves | ||
| Aluminum foil | |||
| Pasteur pipettes | |||
| 1.5 ml microfuge tubes | |||
| Nail polish | clear or colored | ||
| P20 micropipette and plastic tips | |||
| Paperclips | 20 - 25 standard metal paperclips linked to form a chain | ||
| Reagents | |||
| 16% EM grade paraformaldehyde | Electron Microscopy Reagents | ||
| Acetic acid | Sigma | ||
| Liquid nitrogen | |||
| 100% Ethanol, chemical grade | |||
| Commercially synthesized, fluorescently labeled oligos | |||
| Long biotinylated probe | Invitrogen; Alternative steps 2.7.1-2.7.3 | e.g., nick translated and biotinylated with BioNick from Invitrogen | |
| Rhodamine-Avidin | Roche; Alternative steps 2.7.1-2.7.3 | for detection of long biotinylated probe | |
| Hybridization buffer | Recipe above | ||
| 4x SSCT | Recipe above | saline-sodium citrate + Tween | |
| 0.1x SSC | Recipe above | saline-sodium citrate | |
| Blocking solution | Recipe above | ||
| SBT | Recipe above | SSC, bovine serum albumin, Tween | |
| 1x PBT | Recipe above | phosphate-buffered saline + Tween | |
| 1x PBS | phosphate-buffered saline | ||
| Hypotonic solution | 0.5% sodium citrate in H2O | ||
| Formamide | Sigma Aldrich | ||
| Vectashield mounting medium with DAPI | Vector laboratories | ||
Odniesienia
- Blattes, R., Kas, E. Fluorescent in situ hybridization (FISH) on diploid nuclei and mitotic chromosomes from Drosophila melanogaster larval tissues. Cold Spring Harbor Protocols. 2009 (9), (2009).
- Dimitri, P. Fluorescent in situ hybridization with transposable element probes to mitotic chromosomal heterochromatin of Drosophila. Methods in Molecular Biology. 260, 29-39 (2004).
- Pardue, M. L. In situ hybridization to polytene chromosomes in Drosophila using digoxigenin-labeled probes. Cold Spring Harbor Protocols. 2011 (8), 1003-1006 (2011).
- Hoskins, R. A., et al. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biology. 3 (12), (2002).
- Hoskins, R. A., et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 316 (58331), 1625-1628 (2007).
- Treangen, T. J., Salzberg, S. L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nature Reviews Genetics. 13 (1), 36-46 (2012).
- He, B., et al. Mapping the pericentric heterochromatin by comparative genomic hybridization analysis and chromosome deletions in Drosophila melanogaster. Genome Research. 22 (12), 2507-2519 (2012).
- Pimpinelli, S., Bonaccorsi, S., Fanti, L., Gatti, M. Fluorescent in situ hybridization (FISH) of mitotic chromosomes from Drosophila larval brain. Cold Spring Harbor Protocols. 2010 (3), (2010).
- Larracuente, A. M., Noor, M. A., Clark, A. G. Translocation of Y-linked genes to the dot chromosome in Drosophila pseudoobscura. Molecular Biology and Evolution. 27 (7), 1612-1620 (2010).
- Ferree, P. M., Barbash, D. A. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biology. 7 (10), e1000234 (2009).
- Williams, B. C., Karr, T. L., Montgomery, J. M., Goldberg, M. L. The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. The Journal of Cell Biology. 118 (4), 759-773 (1992).
- Gatti, M., Bonaccorsi, S., Pimpinelli, S. Looking at Drosophila Mitotic Chromosomes. Methods in Cell Biology. 44, 371-391 (1994).
- Werren, J. H., Stouthamer, R. PSR (paternal sex ratio) chromosomes: the ultimate selfish genetic elements. Genetica. 117 (1), 85-101 (2003).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone