Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Lens-free Video Microscopy for the Dynamic and Quantitative Analysis of Adherent Cell Culture
W tym Artykule
Podsumowanie
Lens-free video microscopy enables us to monitor cell cultures directly inside the incubator. Here we describe the full protocol used to acquire and analyze a 2.7 day long acquisition of cultured HeLa cells, leading to a dataset of 2.2 x 106 measurements of individual cell morphology and 10584 cell cycle tracks.
Streszczenie
Here, we demonstrate that lens-free video microscopy enables us to simultaneously capture the kinetics of thousands of cells directly inside the incubator and that it is possible to monitor and quantify single cells along several cell cycles. We describe the full protocol used to monitor and quantify a HeLa cell culture for 2.7 days. First, cell culture acquisition is performed with a lens-free video microscope, and then the data is analyzed following a four-step process: multi-wavelength holographic reconstruction, cell-tracking, cell segmentation and cell division detection algorithms. As a result, we show that it is possible to gather a dataset featuring more than 10,000 cell cycle tracks and more than 2 x 106 cell morphological measurements.
Wprowadzenie
Monitoring cultured mammalian cells throughout several cell cycles and measuring accurately cell size and cell dry mass is a challenging task. Several label-free optical techniques are able to perform this task1,2: phase-shifting interferometry3, digital holographic microscopy (DHM)4,5,6,7, quadriwave lateral shearing interferometry8,9 and quantitative phase tomography10,11. These methods have led to many new insights into the understanding of the cell cycle of mammalian cells. However they are seldom coupled with automatic cell tracking algorithms and their throughput remains limited when measuring cell mass trajectories1 (N<20 in respectively3,4,5,6). Hence a novel optical method is needed to measure cell mass trajectories with large statistics (N>1000).
In this paper, we demonstrate the capability of lens-free video microscopy to simultaneously image thousands of cells directly inside the incubator, and then quantify single cell metrics along thousands of individual cell cycle tracks. Lens-free microscopy is a quantitative phase imaging technique which allows the acquisition of phase image of densely packed cells over a very large field of view (typically several tens of mm2, here 29.4 mm2)12,13,14,15. Several metrics at the single cell level are determined, e.g., cell area, cell dry mass, cell thickness, cell major axis length and cell aspect ratio12,15, from each image. Then, by applying a cell-tracking algorithm, these features can be plotted for every single cell as a function of the experiment time14,15. Furthermore, by detecting the occurrence of cell divisions in the cell tracks, it is possible to extract other important information such as the initial cell dry mass (just after the cell division), the final cell dry mass (just before the cell division) and the cell cycle duration, i.e., the time between two consecutive divisions15. All these measurements can be computed with very good statistics (N>1000) since the large field of view would typically allow the analysis of 200 to 10,000 cells in a single lens-free acquisition.
In order to explain this methodology based on lens-free video microscopy, we describe the protocol to monitor and quantify a HeLa cell culture for 2.7 days. Data analysis is a four-step process based on multi-wavelength holographic reconstruction, cell-tracking, cell segmentation and cell division algorithms. Here it is shown that the spatial resolution and the relatively fast frame rate (one acquisition every 10 minutes) obtained with this lens-free video microscopy setup is compatible with standard cell-tracking algorithms. The full analysis of this dataset results in the measurement of 10,584 cell tracks over complete cell cycles.
To summarize, lens-free video microscopy is a powerful tool to automatically monitor thousands of unlabeled, unsynchronized, and unmodified cells per experiment; each cell being tracked over several cell cycles. Our measurements thus provide the mean value of several cell parameters, but more importantly, the inter-cell variability over a large population of cells.
Protokół
1. Cell Culture Monitoring Acquisition
- Grow HeLa cells in DMEM + glutamine (e.g., GlutaMAX) medium supplemented with 10% (v/v) heat-inactivated fetal calf serum and 1% penicillin and streptomycin.
- Coat 6-well glass bottom culture plates with fibronectin (25 µg/mL) for 1 h. Then seed 2 x 104 cells per well.
- During the acquisition, change the medium every 3 days.
- For the time-lapse acquisition, use the video lens-free microscope (commercially available).
NOTE: This is based on the lens-free computational imaging technique as described by Ozcan et al.16 which was modified to perform continuous monitoring in an incubator at a controlled temperature of 37 °C12,15. It features a complementary metal oxide semiconductor (CMOS) image sensor with a pixel pitch of 1.67 µm and an imaging area of 6.4 x 4.6 mm2. Multiple wavelength illumination is provided by a multichip light emitting diode (LEDs) device, which delivers red, green and blue illumination. The wavelengths are centered, on 636, 521, and 452 nm respectively, with a spectral bandwidth of 25, 45, and 25 nm respectively. The red-green-blue (RGB) LEDs are located above a 150 µm pinhole at a distance of approximately 5 cm from the cells. The light power measured close to the LED is as low as 10 µW at each wavelength and the illumination time is only one second per acquisition. Hence, no photo-toxicity problems are expected when cell cultures are observed under the lens-free video microscope. - Put the cell culture container put in contact with the CMOS sensor (Figure 1). Use a container with a glass coverslip at the bottom, which is important for the quality of the lens-free acquisition. Plastic containers can also be used but the lens-free acquisition will be degraded owing to the poor optical quality of these containers.
- Control the video lens-free microscope with the acquisition software (commercially available), which performs both the time-lapse acquisition and the holographic reconstruction. The parameters that need to be entered by the user in the software interface are the frame rate, the duration of the experiment, and the type of cell culture, i.e. adherent cells or floating cells.
- Set the input parameters of the acquisition software. Set the frame rate set to one acquisition every 10 minutes and set the cell culture type to 'adherent'.
NOTE: A frame rate of one acquisition every 10 minutes is a good compromise as it ensures a reliable cell tracking while the size of the resulting dataset to be analyzed is still reasonable. The fastest frame rate achievable with this lens-free microscopy setup is one acquisition every 5 minutes. Further increasing the framerate results in excessive heating of the CMOS sensor which can affect the viability of the cells in culture. - Start the time-lapse acquisition, and the holographic reconstruction algorithm (commercially available) will automatically process the lens-free acquisition to obtain the phase image of the cell culture. The holographic reconstruction algorithm is based on a multi-wavelength phase retrieval algorithm17 and provides an RGB reconstructed phase image in the range of [-π, +π] for each single cell over a large field of view of 29.4 mm2. The principle of lens-free in-line holography is to illuminate a thin sample (at z=0) with a plane wave (of amplitude 1 after normalization) and to detect on a CMOS sensor the subsequent diffraction intensity image at a short distance z=Z, typically 1 mm after the sample. In our setup, the illumination is sequentially switched to 3 wavelengths (λ1=0.450 µm, λ2=0.540 µm, λ3=0.647 µm) to measure three diffraction patterns of the same sample.
2. Cell Culture Data Analysis
- For cell-tracking, use the Trackmate algorithm, an open source Fiji plugin for the automated tracking of single particles18. At first, load the full time-lapse acquisition into Fiji (load image sequence command). Owing to the large size of the datasets, which features typically 400 RGB frames of 29.7 Mb, it is faster to load them in 8-bit format. Next the Trackmate Fiji plugin guides the user through several stages of the cell-tracking algorithm, namely a cell detection stage, a cell tracker stage and several filters applied to the cell detections and the computed tracks. At every stage, configure the algorithms and display the results immediately so that, if necessary, one can easily navigate back and forth to readjust the settings. The various menus of the Trackmate interface are shown in Figure 2 with the actual settings used for the HeLa cells experiment.
- Set the input parameters of the acquisition software as following (see Figure 2). Set the estimated blob diameter to 15 pixels, the detector threshold to 0.25, the linking maximum distance to 15 pixels, the gap-closing max distance to 15 pixels, and the number of spots in tracks to 3.5.
NOTE: These settings will obviously differ from one experiment to another, especially the 'estimated blob diameter' which corresponds to the cell size (here given in pixels of 1.67 µm). The same remark applies to the 'linking max distance' which account for the maximum distance travelled by a cell within two consecutive frames (here given in pixels of 1.67 µm). Its optimal value would depend on cell motility but also on cell density. - At the end of the cell-tracking process, generate the results in the form of three text files ('Analysis' button, see Figure 2). The most useful file, named 'Spots in tracks statistics', consists of a table listing all detected cells with their respective (x,y) positions in the acquisition, their frame number and their track number. On the basis of this data, perform cell segmentation and detect cell divisions using dedicated algorithms (see supplementary code files). The two other files are named 'Links in tracks statistics' and 'Track statistics' and include all the results dealing with the tracks, e.g. track durations, number of detected gaps, track initial frame, etc.
- Use the cell segmentation algorithm (see supplementary code file) to automatically extract several metrics describing the cell morphology from the reconstructed phase image. These metrics are the cell surface area (S), the cell major axis length (L), the cell minor axis length (l) and the cell aspect ratio (L/l).The phase recovered from lens-free microscopy is proportional to the density and thickness of the specimen layer as discussed previously15.
- In addition to the morphological features of the cell, extract quantitative cell dry mass (CDM) measurements from the reconstructed phase13,19,20
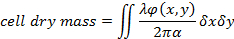 Eq. (1)
Eq. (1)
where φ(x,y) is the reconstructed phase shift19, λ the wavelength and α = 1.8 x 10-4 m3/kg the specific refractive index which relates the variation of refractive index to dry mass1,19. - Evaluate the cell thickness using an estimation of the difference between the cell refractive index and that of the culture media. The relation between the cell thickness h(x,y) and the measured phase shift is given by20:
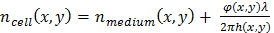 Eq. (2.1)
Eq. (2.1)
 Eq. (2.2)
Eq. (2.2)
with Δn(x,y) = ncell (x,y) - nmedium (x,y), the refractive index difference between the cell and the culture media. In the following, assume a constant value of Δn = 0.025, which is estimated from the refractive index measured in HeLa cell nuclei (n = 1.35511) and that of phosphate-buffered saline (PBS) culture media (n=1.33). Although the cell thickness value is only indicative as it is based on an assumption, its relative variations are meaningful and can be exploited to detect dividing cells. - Use the dedicated algorithm (see supplementary code file) to detect the occurrence of cell divisions and then extract cell tracks that are associated to the detected cell divisions, In order to detect a cell division, cells with a measured thickness larger than 8 µm are first identified, then check whether a new cell appears closely in space and time. In this way, the detection of cell divisions relies on two robust criteria. Alternative and more elaborate methods for cell division detection can be applied to the lensfree time-lapse acquisition21,22,23.
Wyniki
For the holographic reconstruction process, the light field is described by a scalar field A (where 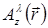 is the complex value of A on the plane at distance z from the sample, and lateral position
is the complex value of A on the plane at distance z from the sample, and lateral position 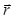 and at wavelength λ). Light propagation is modeled by the Huygens-Fresnel theory which provides a propagator kernel
and at wavelength λ). Light propagation is modeled by the Huygens-Fresnel theory which provides a propagator kernel
Dyskusje
In this paper, we show that lens-free video microscopy can be used inside an incubator to capture the kinetics of thousands of cells. In order to describe the overall methodology we explained how a 2.7 day time-lapse acquisition of HeLa cells in culture can be analyzed with standard cell-tracking algorithms. The result is a dataset featuring 2.2 x 106 cell measurements and 10,584 cell cycle tracks. The acquisitions were performed on a culture of Hela cells with a relatively large cell-to-cell distance (cell de...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
The authors have nothing to acknowledge.
Materiały
| Name | Company | Catalog Number | Comments |
| Cytonote lens-free video microscope | Iprasense | ||
| Horus acquisition software | Iprasense | ||
| 6-well glass bottom culture plates | MatTek corporation | Part No: P06G-0-14-F | |
| DMEM + GlutaMAX medium | Gibco | ||
| heat-inactivated fetal calf serum | Eurobio | ||
| penicillin and streptomycin | Gibco | ||
| Fibronectin | Sigma Aldrich | ||
| Matlab, image processing toolbox | Mathworks |
Odniesienia
- Zangle, T. A., Teitell, M. A. Live-cell mass profiling: an emerging approach in quantitative biophysics. Nat Methods. 11 (12), 1221-1228 (2014).
- Popescu, G., Park, K., Mir, M., Bashir, R. New technologies for measuring single cell mass. Lab Chip. 14 (4), 646-652 (2014).
- Reed, J., et al. Rapid, massively parallel single-cell drug response measurements via live cell interferometry. Biophys J. 101 (5), 1025-1031 (2011).
- Mir, M., et al. Optical measurement of cycle-dependent cell growth. Proc Natl Acad Sci. USA. 108 (32), 13124-13129 (2011).
- Girshovitz, P., Shaked, N. T. Generalized cell morphological parameters based on interferometric phase microscopy and their application to cell life cycle characterization. Biomed Opt Express. 3 (8), 1757-1773 (2012).
- Kemper, B., Bauwens, A., Vollmer, A., Ketelhut, S., Langehanenberg, P. Label-free quantitative cell division monitoring of endothelial cells by digital holographic microscopy. J Biomed Opt. 15 (3), (2010).
- Mir, M., Bergamaschi, A., Katzenellenbogen, B. S., Popescu, G. Highly sensitive quantitative imaging for monitoring single cancer cell growth kinetics and drug response. PLoS One. 9 (2), 1-8 (2014).
- Bon, P., Savatier, J., Merlin, M., Wattellier, B., Monneret, S. Optical detection and measurement of living cell morphometric features with single-shot quantitative phase microscopy. J Biomed Opt. 17 (7), (2012).
- Aknoun, S., et al. Living cell dry mass measurement usinq quantitative phase imaging with quadriwave lateral shearing interferometry: an accuracy and sensitivity discussion. J Biomed Opt. 20 (1), 1-4 (2015).
- Cotte, Y., et al. Marker-free phase nanoscopy. Nature Photonics. 7 (2), 113-117 (2013).
- Choi, W., et al. Tomographic phase microscopy. Nat Methods. 4 (9), 717-719 (2007).
- Kesavan, S. V., et al. High-throughput monitoring of major cell functions by means of lensfree video microscopy. Sci Rep. 4, 1-11 (2014).
- Zheng, G., Lee, S. A., Antebi, Y., Elowitz, M. B., Yang, C. The ePetri dish, an on-chip cell imaging platform based on subpixel perspective sweeping microscopy (SPSM). Proc Natl Acad Sci USA. 108 (41), 16889-16894 (2011).
- Pushkarsky, I., et al. Automated single-cell motility analysis on a chip using lensfree microscopy. Sci Rep. 4, 4717 (2014).
- Allier, C., et al. Imaging of dense cell cultures by multiwavelength lens-free video microscopy. Cytom Part A. 91 (5), 1-10 (2017).
- Su, T. -. W., Seo, S., Erlinger, A., Ozcan, A. High-throughput lensfree imaging and characterization of a heterogeneous cell solution on a chip. Biotechnol Bioeng. 102 (3), 856-868 (2009).
- Delacroix, R., et al. Cerebrospinal fluid lens-free microscopy: a new tool for the laboratory diagnosis of meningitis. Sci Rep. 7, 39893 (2017).
- Tinevez, J. -. Y., et al. TrackMate: an open and extensible platform for single-particle tracking. Methods. 115, 80-90 (2016).
- Popescu, G. Optical imaging of cell mass and growth dynamics. Am J Physiol Physiol. 295 (2), 538-544 (2008).
- Liu, P. Y., et al. Cell refractive index for cell biology and disease diagnosis: past, present and future. Lab Chip. 16, 634-644 (2016).
- Rapoport, D. H., Becker, T., Mamlouk, A. M., Schicktanz, S., Kruse, C. A novel validation algorithm allows for automated cell tracking and the extraction of biologically meaningful parameters. PLoS One. 6 (11), e27315 (2011).
- Al-Kofahi, O., et al. Automated cell lineage construction: A rapid method to analyze clonal development established with murine neural progenitor cells. Cell Cycle. 5 (3), 327-335 (2006).
- Meijering, E., Dzyubachyk, O., Smal, I., van Cappellen, W. A. Tracking in cell and developmental biology. Semin Cell Dev Biol. 20 (8), 894-902 (2009).
- Posakony, J. W., England, J. M., Attardi, G. Mitochondrial growth and division during the cell cycle in HeLa cells. J Cell Biol. 74 (2), 468-491 (1977).
- Zocchi, E., et al. Expression of CD38 Increases Intracellular Calcium Concentration and Reduces Doubling Time in HeLa and 3T3 Cells. J Biol Chem. 273 (14), 8017-8024 (1979).
- Reitzer, L. J., Wice, B. M., Kennell, D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 254 (8), 2669-2676 (1979).
- Benedetti, A. D. E., Joshi-barve, S., Rinker-Schaeffer, C., Rhoads, R. E. Expression of Antisense RNA against Initiation Factor eIF-4E mRNA in HeLa Cells Results in Lengthened Cell Division Times, Diminished Translation Rates, and Reduced Levels of Both eIF-4E and the p220 Component of eIF-4F. Mol Cell Biol. 11 (11), 5435-5445 (1991).
- Kumei, Y., Nakajima, T., Sato, A., Kamata, N., Enomoto, S. Reduction of G1 phase duration and enhancement of c-myc gene expression in HeLa cells at hypergravity. J Cell Sci. 93 (2), 221-226 (1989).
- Ginzberg, M. B., Kafri, R., Kirschner, M. On being the right (cell) size. Science. 348 (6236), 1245075 (2015).
- Mathieu, E., et al. Time-lapse lens-free imaging of cell migration in diverse physical microenvironments. Lab Chip. 16 (17), 3304-3316 (2016).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone