Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Polysome Purification from Soybean Symbiotic Nodules
W tym Artykule
Podsumowanie
This protocol describes a method for eukaryotic polysome purification from intact soybean nodules. After sequencing, standard pipelines for gene expression analysis can be used to identify differentially expressed genes at the transcriptome and translatome levels.
Streszczenie
The aim of this protocol is to provide a strategy for studying the eukaryotic translatome of the soybean (Glycine max) symbiotic nodule. This paper describes methods optimized to isolate plant-derived polyribosomes and their associated mRNAs to be analyzed using RNA-sequencing. First, cytoplasmic lysates are obtained through homogenization in polysome- and RNA-preserving conditions from whole, frozen soybean nodules. Then, lysates are cleared by low-speed centrifugation, and 15% of the supernatant is used for total RNA (TOTAL) isolation. The remaining cleared lysate is used to isolate polysomes by ultracentrifugation through a two-layer sucrose cushion (12% and 33.5%). Polysome-associated mRNA (PAR) is purified from polysomal pellets after resuspension. Both TOTAL and PAR are evaluated by highly sensitive capillary electrophoresis to meet the quality standards of sequencing libraries for RNA-seq. As an example of a downstream application, after sequencing, standard pipelines for gene expression analysis can be used to obtain differentially expressed genes at the transcriptome and translatome levels. In summary, this method, in combination with RNA-seq, allows the study of the translational regulation of eukaryotic mRNAs in a complex tissue such as the symbiotic nodule.
Wprowadzenie
Leguminous plants, such as soybean (Glycine max), can establish symbiosis with specific soil bacteria called rhizobia. This mutualistic relationship elicits the formation of novel organs, the symbiotic nodules, on the plant roots. The nodules are the plant organs hosting the bacteria and consist of host cells whose cytoplasm is colonized with a specialized form of rhizobia called bacteroids. These bacteroids catalyze the reduction of atmospheric nitrogen (N2) into ammonia, which is transferred to the plant in return for carbohydrates1,2.
Although this nitrogen-fixing symbiosis is one of the most well-studied plant-microbe symbioses, many aspects remain to be better understood, such as how plants subjected to different abiotic stress conditions modulate their interaction with their symbiotic partner and how this affects nodule metabolism. These processes could be better understood by analyzing the nodule translatome (i.e., the subset of messenger RNAs [mRNAs] actively translated). Polyribosomes or polysomes are complexes of multiple ribosomes associated with mRNA, commonly used to study translation3. The polysome profiling method consists of the analysis of the mRNAs associated with polysomes and has been successfully used to study the posttranscriptional mechanisms controlling gene expression that occurs in diverse biological processes4,5.
Historically, genome expression analysis has focused primarily on determining mRNA abundance6,7,8,9. However, there is a lack of correlation between transcript and protein levels due to the different stages of posttranscriptional regulation of gene expression, particularly translation10,11,12. Moreover, no dependence has been observed between the changes at the level of the transcriptome and those that occur at the level of the translatome13. The direct analysis of the set of mRNAs that are being translated allows a more accurate and complete measurement of the cell gene expression (whose endpoint is protein abundance) than the one obtained when only mRNA levels are analyzed14,15,16.
This protocol describes how plant-derived polysomes are purified from intact soybean nodules by differential centrifugation through a two-layer sucrose cushion (Figure 1). However, since bacteroid-derived ribosomes are also present in the nodules, a mix of ribosomes and RNA species are purified, even though the eukaryotic ones represent the main fraction (90%-95%). The subsequent RNA isolation, quantification, and quality control are also described (Figure 1). This protocol, in combination with RNA-seq, should provide experimental results on the translational regulation of eukaryotic mRNAs in a complex tissue such as the symbiotic nodule.
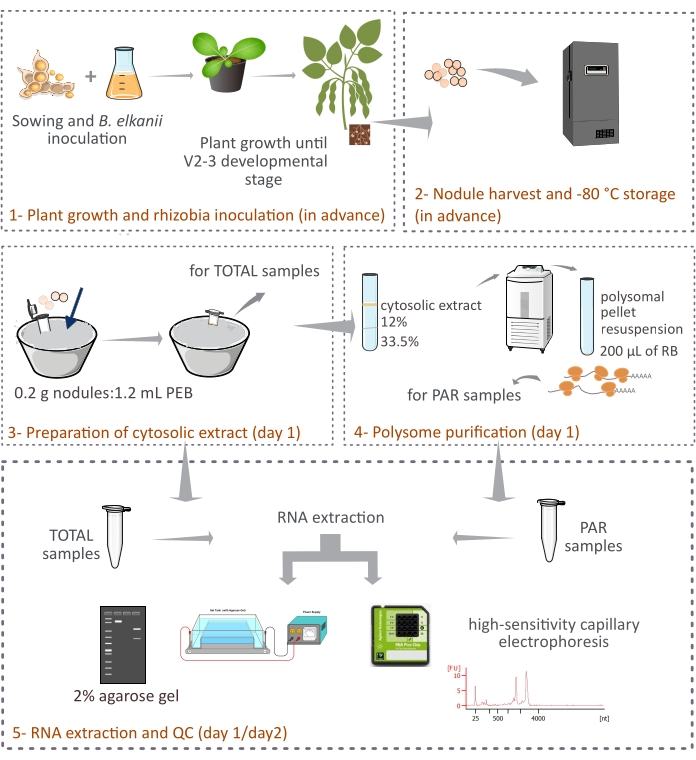
Figure 1: Schematic overview of the proposed methodology for eukaryotic polysome purification from symbiotic nodules. The scheme gives an overview of the steps followed in the protocol from (1) plant growth and (2) nodule harvest to (3) preparation of the cytosolic extracts, (3) obtaining TOTAL samples and (4) PAR samples, and (5) RNA extraction and quality control. Abbreviations: PEB = polysome extraction buffer; RB = resuspension buffer; TOTAL = total RNA; PAR = polysome-associated mRNA. Please click here to view a larger version of this figure.
Protokół
1. Plant growth and rhizobia inoculation
- To generate the required nodules, sow the soybean seeds of choice in the selected substrate in a growth chamber under controlled conditions.
NOTE: In this protocol, seeds were sown in a 0.5 L plastic bottle filled with a mix of sand:vermiculite (1:1). The growth chamber was set with a day/night cycle temperature of 28 °C /20 °C, respectively, and a light/darkness photoperiod of 16 h/8 h, respectively. The photosynthetically active radiation intensity was 620 µmol·m−2·s−1. - Prepare in advance liquid Yeast-Extract Mannitol (YEM)-medium (see the Table of Materials). Autoclave the medium.
- On the same day of seed sowing, inoculate one flask containing 100 mL of YEM with the Bradyrhizobium elkanii U1302 strain. Incubate the flask at 28 °C on an orbital shaker at 100 rpm.
- Let the rhizobia grow for 2 days and inoculate the seedlings with 2 mL of the culture.
2. Water deficit treatment (optional)
NOTE: This protocol outlines the water deficit treatment of the soybean plants. This part can be changed or omitted entirely depending on the experimental question at hand.
- Grow the soybean seedlings for 19 days (V2-3 developmental stage) without water restriction. Keep the substrate at field capacity with B&D-medium17 supplemented with 0.5 mM KNO3.
- On day 20, withdraw watering.
NOTE: Here, the water content was measured daily by gravimetry (water gravimetric content) during the growth and water deficit periods. - Measure stomatal conductance in triplicate daily on the abaxial leaf surface with a porometer, as instructed by the manufacturer (see the Table of Materials), from day 20 until the end of the water deficit period.
NOTE: The end of the water deficit period was determined individually for each plant when the stomatal conductance value was approximately 50% of the one obtained on day 20 from sowing.
3. Nodule harvest
- Collect liquid nitrogen in a clean container and label 15 mL tubes (one for each plant).
CAUTION: Handle liquid nitrogen wearing cryo gloves, face shields, and safety goggles. - Retrieve each plant individually. Cut and discard the aerial part. Thoroughly wash the root with water to remove any remaining substrate.
- Detach each nodule from the root and collect them in a prechilled 15 mL tube. Snap-freeze the tubes and store them at −80 °C.
4. Preparation of cytosolic extracts
NOTE: The final aim of this protocol is to obtain high-quality total RNA (TOTAL) and polysome-associated RNA (PAR). Therefore, work under conditions that prevent RNA degradation, always keeping the samples at 4 °C and using RNase-free laboratory equipment and solutions. Unless specified, all the solutions are prepared with sterile ultrapure water.
- Preparation of buffer stock solutions
- Prepare the autoclavable stock solutions listed in Table 1, autoclave them for 15 min, and keep at RT.
- Prepare the filter-sterilizable stock solutions listed in Table 1, filter-sterilize them, and keep them at −20 °C.
- Prepare polysome extraction buffer (PEB, see Table 1) and keep it on ice.
NOTE: The volumes given are for six samples. - Cool the centrifuge to 4 °C and place 2 mL microcentrifuge tubes on ice.
- Weigh approximately 0.2 g of intact nodules in a weighing dish and transfer them to a precooled 2 mL tube.
NOTE: Perform this step quickly to avoid the thawing of the samples. Alternatively, an estimated nodule volume of 0.4-0.5 mL can be used instead of the 0.2 g. - Add 1.2 mL of PEB, let the sample thaw for 2 min, and homogenize with a tissue grinder until complete disruption and homogenization of the nodules.
NOTE: Be sure to grind the nodules once thawed as they will be soft enough to be easily disrupted with tissue grinders (see the Table of Materials) in microcentrifuge tubes. Frozen nodules are difficult to grind. Also, it is recommended to place the tubes in a benchtop cooler (0 °C) for proper homogenization while keeping samples cool. - Incubate the samples on ice with gentle agitation for 10 min or until all samples have been processed.
- Centrifuge at 16,000 × g for 15 min at 4 °C to pellet the debris. Recover the supernatant and repeat the centrifuge step.
- Carefully recover the clarified cytosolic extract and transfer a 200 µL aliquot to a clean 1.5 mL microcentrifuge tube for TOTAL isolation (TOTAL samples; see section 7).
5. Preparation of sucrose cushions
NOTE: This protocol uses a two-layer sucrose cushion (12% and 33.5%) in 13.2 mL ultracentrifuge tubes (see the Table of Materials). All solutions are prepared with sterile ultrapure water.
- Prepare the sucrose and salt stock solutions.
- Prepare 100 mL of 2 M sucrose solution (68.5%, Table 1).
- Prepare 10x salt solution (Table 1).
- Prepare the two layers of the sucrose cushion as described in Table 2. See Table 1 for the composition of the antibiotic (CHX and CHL) stock solutions.
NOTE: The volumes given are for six cushions. - Pour 4.5 mL of the 33.5% sucrose layer into the centrifuge tubes. Then, add 4.5 mL of the 12% layer carefully and slowly with a P1000 micropipette. Put the tubes on ice until the addition of the clarified cytosolic extract.
6. Polysome purification
- Load 1 mL of the clarified cytosolic extract (step 4.8) on top of the sucrose cushion by carefully pipetting onto the sidewall of the tube.
- Transfer the ultracentrifuge tubes to precooled buckets and centrifuge at 217,874 × g (35,000 rpm in the referenced rotor [see the Table of Materials]) for 2 h at 4 °C.
- Discard the remaining cytosolic extract and sucrose cushion, and resuspend the polysomal pellet with 200 µL of precooled RB (previously prepared according to Table 1).
- Incubate for 30 min on ice and then transfer the polysomal resuspension to 1.5 mL precooled tubes. Proceed with the RNA extraction (PAR samples).
7. RNA extraction and quality control
NOTE: This step is performed for TOTAL (step 4.8) and PAR samples (step 6.4).
- Prepare 75% EtOH with sterile ultrapure water and cool it to −20 °C. Additionally, cool chloroform and isopropanol to −20 °C.
- Homogenize the samples with 750 µL of the RNA isolation reagent and incubate for 5 min at RT.
- Add 200 µL of cold chloroform and shake the tubes vigorously for 15 s. Incubate at RT for 10 min.
- Centrifuge at 12,000 × g for 15 min at 4 °C for phase separation. Transfer 500 µL of the upper aqueous phase to a clean tube without disturbing the pink organic phase, and add 375 µL of cold isopropanol and 0.5 µL of RNase-free glycogen (see the Table of Materials). Mix thoroughly by pipetting up and down.
NOTE: Glycogen is a carrier used as a co-precipitant to increase nucleic acid recovery from alcohol precipitation. - Incubate the mix for 10 min at 4 °C and centrifuge at 12,000 × g for 15 min. Discard the supernatant.
- Wash the RNA precipitate with 1 mL of cold 75% EtOH. Mix by brief vortexing.
NOTE: At this point, the samples can be stored at −20 °C overnight. - Centrifuge at 7,500 × g for 5 min at 4 °C, carefully remove the supernatant with a micropipette, and air-dry the RNA pellet.
- Dissolve the pellet in 50 µL of RNase-free water and incubate at 65 °C for 5 min.
- Assess the RNA concentration and integrity with highly sensitive capillary electrophoresis18 (see the Table of Materials) and/or electrophoresis on a 2% RNase-free agarose gel19 (see the Table of Materials).
NOTE: If the RNA samples are not going to be used immediately or are going to be sent to the sequencing facility for RNA-seq library preparation, it is recommended to use EtOH to precipitate them.
8. RNA precipitation
- Cool the centrifuge and EtOH to 4 °C.
- Prepare 3 M sodium acetate.
- Prepare 70% EtOH with sterile ultrapure water and cool it to −20 °C.
- Estimate the sample volume. Add 0.1 volumes of 3 M sodium acetate, 3 volumes of cold EtOH, and 0.5 µL of RNase-free glycogen. Mix thoroughly.
- Leave at −20 °C until needed.
NOTE: RNA precipitated samples can be stored for up to 1 year at −80 °C.
9. Standard pipeline for gene expression analysis
- Perform an average of 40M Illumina paired-end reads, with read length >100 bp, for confident transcriptome and translatome data analysis.
NOTE: Standard RNA-seq data analysis includes the following steps 9.2-9.5. - Carry out read quality inspection using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) or the multisample MultiQC20.
- Remove adaptor and low-quality sequences using either Trimmomatic21, BBDuk (http://jgi.doe.gov/data-and-tools/bb-tools/), cutadapt22, sickle23, or Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), among others.
- If a reference genome is available for the species, carry out read mapping against the reference, followed by abundance quantification, using Bowtie224, TopHat225 or Salmon26, and featureCounts27, besides other open source tools.
- Perform statistical analysis for differential expressed genes identification using edgeR28, DESeq229, or limma30, among others.
NOTE: These open-source software can be used locally, via the command line. Alternatively, they can be operated through a web browser on a public server, such as Galaxy31 or GEOexplorer32, which offer a graphical user interface, so that no command line knowledge is required for their use.
Wyniki
The quantity and quality assessment of the TOTAL and PAR fractions purified with the abovementioned procedure is key to determining its success, since for most downstream applications, such as RNA sequencing, high-quality samples are fundamental for library preparation and sequencing. Moreover, the integrity of the RNA molecules allows the capture of a snapshot of the gene expression profile at the moment of sample collection18. In this context, an RNA integrity number (RIN) is obtained when perfo...
Dyskusje
Studying gene expression regulation at the translational level is critical to better comprehend different biological processes since the endpoint of cell gene expression is protein abundance13,14. This can be assessed by analyzing the translatome of the tissue or organism of interest for which the polysomal fraction should be purified and its associated mRNAs analyzed3,4,34
Ujawnienia
The authors have no conflicts of interest.
Podziękowania
This research was funded by CSIC I+D 2020 grant No. 282, FVF 2017 grant No. 210, and PEDECIBA (María Martha Sainz).
Materiały
| Name | Company | Catalog Number | Comments |
| Plant growth and rhizobia inoculation | |||
| Orbital shaker | Daihan Scientific | Model SHO-1D | |
| YEM-medium | Amresco | J850 (yeast extract) 0122 (mannitol) | |
| Water deficit treatment | |||
| KNO3 | Merck | 221295 | |
| Porometer | Decagon Device | Model SC-1 | |
| Scalpel | |||
| Preparation of cytosolic extracts | |||
| Brij L23 | Sigma-Aldrich | P1254 | |
| Centrifuge | Sigma | Model 2K15 | |
| Chloranphenicol | Sigma-Aldrich | C0378 | |
| Cycloheximide | Sigma-Aldrich | C7698 | |
| DOC | Sigma-Aldrich | 30970 | |
| DTT | Sigma-Aldrich | D9779 | |
| EGTA | Sigma-Aldrich | E3889 | |
| Igepal CA 360 | Sigma-Aldrich | I8896 | |
| KCl | Merck | 1.04936 | |
| MgCl2 | Sigma-Aldrich | M8266 | |
| Plastic tissue grinder | Fisher Scientific | 12649595 | |
| PMSF | Sigma-Aldrich | P7626 | |
| PTE | Sigma-Aldrich | P2393 | |
| Tris | Invitrogen | 15504-020 | |
| Triton X-100 | Sigma-Aldrich | T8787 | |
| Tween 20 | Sigma-Aldrich | P1379 | |
| Weighing dish | Deltalab | 1911103 | |
| Preparation of sucrose cushions | |||
| Sucrose | Invitrogen | 15503022 | |
| SW 40 Ti rotor | Beckman-Coulter | ||
| Ultracentrifuge | Beckman-Coulter | Optima L-100K | |
| Ultracentrifuge tubes | Beckman-Coulter | 344059 | 13.2 mL tubes |
| RNA extraction and quality control | |||
| Agarose | Thermo scientific | R0492 | |
| Bioanalyzer | Agilent | Model 2100. Eukaryote total RNA nano assay | |
| Chloroform | DI | 41191 | |
| Ethanol | Dorwil | UN1170 | |
| Isopropanol | Mallinckrodt | 3032-06 | |
| Glycogen | Sigma | 10814-010 | |
| TRIzol LS | Ambion | 102960028 | |
| Miscellaneous | |||
| Falcon tubes 15 mL | Biologix | 10-0152 | |
| Filter tips 10 µL | BioPointe Scientific | 321-4050 | |
| Filter tips 1000 µL | BioPointe Scientific | 361-1050 | |
| Filter tips 20 µL | BioPointe Scientific | 341-4050 | |
| Filter tips 200 µL | Tarsons | 528104 | |
| Microcentrifuge tubes 1.5 mL | Tarsons | 500010-N | |
| Microcentrifuge tubes 2.0 mL | Tarsons | 500020-N | |
| Sequencing company | Macrogen | ||
| Sterile 250 mL flask | Marienfeld | 4110207 |
Odniesienia
- Limpens, E., et al. Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS ONE. 8 (5), 64377 (2013).
- Masson-Boivin, C., Giraud, E., Perret, X., Batut, J. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes. Trends in Microbiology. 17 (10), 458-466 (2009).
- King, H. A., Gerber, A. P. Translatome profiling: Methods for genome-scale analysis of mRNA translation. Briefings in Functional Genomics. 15 (1), 22-31 (2014).
- Chassé, H., Boulben, S., Costache, V., Cormier, P., Morales, J. Analysis of translation using polysome profiling. Nucleic Acids Research. 45 (3), 15 (2017).
- Yángüez, E., Castro-Sanz, A. B., Fernández-Bautista, N., Oliveros, J. C., Castellano, M. M. Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PloS ONE. 8 (8), 71425 (2013).
- Brown, P. O., Botstein, D. Exploring the new world of the genome with DNA microarrays. Nature Genetics. 21, 33-37 (1999).
- Krishnamurthy, A., Ferl, R. J., Paul, A. -. L. Comparing RNA-Seq and microarray gene expression data in two zones of the Arabidopsis root apex relevant to spaceflight. Applications in Plant Sciences. 6 (11), 1197 (2018).
- Shulse, C. N., et al. High-throughput single-cell transcriptome profiling of plant cell types. Cell Reports. 27 (7), 2241-2247 (2019).
- Wang, Z., Gerstein, M., Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nature Reviews Genetics. 10 (1), 57-63 (2009).
- Kawaguchi, R., Girke, T., Bray, E. A., Bailey-Serres, J. Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. The Plant Journal. 38 (5), 823-839 (2004).
- Larsson, O., Tian, B., Sonenberg, N. Toward a genome-wide landscape of translational control. Cold Spring Harbor Perspectives in Biology. 5 (1), 012302 (2013).
- Vogel, C., Marcotte, E. M. Insights into regulation of protein abundance from proteomics and transcriptomis analyses. Nature Reviews Genetics. 13 (4), 227-232 (2013).
- Tebaldi, T., et al. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics. 13 (220), 1-15 (2012).
- Wang, T., et al. Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic Acids Research. 41 (9), 4743-4754 (2013).
- Ingolia, N. T. Ribosome profiling: New views of translation, from single codons to genome scale. Nature Reviews Genetics. 15 (3), 205-213 (2014).
- Lukoszek, R., Feist, P., Ignatova, Z. Insights into the adaptive response of Arabidopsis thaliana to prolonged thermal stress by ribosomal profiling and RNA-Seq. BMC Plant Biology. 16 (1), 1-13 (2016).
- Broughton, W. J., Dilworth, M. J. Control of leghaemoglobin synthesis in snake beans. Biochemical Journal. 125 (4), 1075-1080 (1971).
- Schroeder, A., et al. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology. 7, 1-14 (2006).
- Rio, D. C., Ares, M., Hannon, G. J., Nilsen, T. W. Nondenaturing agarose gel electrophoresis of RNA nondenaturing agarose gel electrophoresis of RNA. Cold Spring Harbor Protocols. 2010 (6), 1-3 (2012).
- Ewels, P., Magnusson, M., Lundin, S., Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 32 (19), 3047-3048 (2016).
- Bolger, A. M., Lohse, M., Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 30 (15), 2114-2120 (2014).
- Marcel, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 17 (1), 10-12 (2011).
- . Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) Available from: https://github.com/najoshi/sickle (2011)
- Langmead, B., Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 9 (4), 357-359 (2012).
- Kim, D., et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 14 (4), 1-13 (2013).
- Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nature Methods. 14 (4), 417-419 (2017).
- Liao, Y., Smyth, G. K., Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30 (7), 923-930 (2013).
- Robinson, M. D., McCarthy, D. J., Smyth, G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26 (1), 139-140 (2010).
- Love, M. I., Huber, W., Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 15 (550), 1-21 (2014).
- Ritchie, M. E., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 43 (7), 47 (2015).
- Afgan, E., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research. 46, 537-544 (2018).
- Hunt, G. P., et al. GEOexplorer: A webserver for gene expression analysis and visualisation. Nucleic Acids Research. , (2022).
- Imbeaud, S., et al. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Research. 33 (6), 1-12 (2005).
- Di Paolo, A., et al. PDCD4 regulates axonal growth by translational repression of neurite growth-related genes and is modulated during nerve injury responses. RNA. 26 (11), 1637-1653 (2020).
- Smircich, P., et al. Ribosome profiling reveals translation control as a key mechanism generating differential gene expression in Trypanosoma cruzi. BMC Genomics. 16 (1), 443 (2015).
- Eastman, G., Sharlow, E. R., Lazo, J. S., Bloom, G. S., Sotelo-Silveira, J. R. Transcriptome and translatome regulation of pathogenesis in Alzheimer's disease model mice. Journal of Alzheimer's Disease. 86 (1), 365-386 (2022).
- Zanetti, M. E., Chang, I. -. F., Gong, F., Galbraith, D. W., Bailey-Serres, J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant physiology. 138 (2), 624-635 (2005).
- Mustroph, A., et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 106 (44), 18843-11848 (2009).
- Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S., Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324 (5924), 218-223 (2009).
- Brar, G. A., Weissman, J. S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nature Reviews. Molecular Cell Biology. 16 (11), 651-664 (2015).
- Eastman, G., Smircich, P., Sotelo-Silveira, J. R. Following ribosome footprints to understand translation at a genome wide level. Computational and Structural Biotechnology Journal. 16, 167-176 (2018).
- Westermann, A. J., Gorski, S. A., Vogel, J. Dual RNA-seq of pathogen and host. Nature Reviews Microbiology. 10 (9), 618-630 (2012).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone