Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Methodology to Metabolically Inactivate Bacteria for Caenorhabditis elegans Research
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
The food source for Caenorhabditis elegans in the lab is live Escherichia coli. Since bacteria are metabolically active, they present a confounding variable in metabolic and drug studies in C. elegans. A detailed protocol to metabolically inactivate bacteria using paraformaldehyde is described here.
Streszczenie
Caenorhabditis elegans is a common model organism for research in genetics, development, aging, metabolism, and behavior. Because C. elegans consume a diet of live bacteria, the metabolic activity of their food source can confound experiments looking for the direct effects of various interventions on the worm. To avoid the confounding effects of bacterial metabolism, C. elegans researchers have used multiple methods to metabolically inactivate bacteria, including ultraviolet (UV)-irradiation, heat-killing, and antibiotics. UV treatment is relatively low-throughput and cannot be used in liquid culture because each plate must be examined for successful bacterial killing. A second treatment method, heat-killing, negatively affects the texture and nutritional quality of the bacteria, leading to the developmental arrest of C. elegans. Finally, antibiotic treatment can directly alter C. elegans physiology in addition to preventing bacterial growth. This manuscript describes an alternative method to metabolically inactivate bacteria using paraformaldehyde (PFA). PFA treatment cross-links proteins within bacterial cells to prevent metabolic activity while preserving cellular structure and nutritional content. This method is high-throughput and can be used in liquid culture or solid plates, as testing one plate of PFA-treated bacteria for growth validates the whole batch. Metabolic inactivation through PFA treatment can be used to eliminate the confounding effects of bacterial metabolism on studies of drug or metabolite supplementation, stress resistance, metabolomics, and behavior in C. elegans.
Wprowadzenie
Caenorhabditis elegans was originally proposed as a model organism in 19651 and has since been widely adopted in studies of genetics, development, behavior, aging, and metabolism2. Due to their large brood size and transparent cuticle, C. elegans is particularly well-suited for high-throughput screening with fluorescent reporters3. Their short life cycle, hermaphroditic reproduction, and genetic homology with humans also make C. elegans a valuable model system for studies on development4 and aging biology5. Moreover, C. elegans are relatively easy to maintain. Worms can be grown in liquid culture or on solid agar plates and consume a diet of live Escherichia coli OP50 bacteria4.
However, the live food source of C. elegans can confound studies of metabolism, drug supplementation, and behavior. Because live bacteria have their own metabolism, experimental conditions that affect the bacteria also alter the nutrients and metabolites available to the worms. For example, differences in bacterial iron, amino acid, and folate concentrations have diverse effects on C. elegans' development, physiology, and lifespan6. Many common lab practices can elicit such changes in the nutrient composition and metabolites produced by OP50. Specifically, exposure to 5-fluoro-2'-deoxyuridine (FUdR), a compound commonly used to prevent reproduction in C. elegans, elicits broad changes in OP50 gene expression, including amino acid biosynthesis pathways7. Live bacteria can also confound studies in which C. elegans are supplemented with small molecules because bacteria can partially or completely metabolize the active compounds. Moreover, the effects of these small molecules on the bacteria can, in turn, alter C. elegans physiology, as was reported with the lifespan-extending drug metformin8. Finally, live bacteria can change the worm's environment in ways that alter behavior, such as secreting attractive odorants9, producing exogenous neuromodulators10, and creating oxygen gradients in a dense bacteria lawn11.
To mitigate the confounding effects of bacterial metabolism on C. elegans research, multiple methods for killing bacteria have been developed (Table 1). Three common strategies for killing OP50 are UV-irradiation, heat-killing, and antibiotic treatment. While straightforward and relatively low-cost, each of these methods can have undesirable effects on both bacteria and C. elegans. UV-killing via a UV crosslinker12 is low-throughput and the rate is limited by the number of plates that can fit in the UV crosslinker. Moreover, the efficacy of UV-killing can vary from plate to plate within a batch, and testing for growth on all plates can become difficult in large experiments. Heat-killing OP50 by exposing culture to temperatures of >60 °C comes with a separate set of challenges. High heat can damage nutrients essential for the worm and destroy the cellular structure of bacteria, creating a softer texture that decreases the amount of time worms spend on the food13. This method also cannot be used throughout the life cycle of C. elegans because worms fed heat-killed bacteria can arrest early in development13. Antibiotic treatment is a third common method for suppressing bacterial metabolism14, but antibiotics can also alter worm growth and metabolism15.
One solution to eliminate the metabolic effects of live bacteria while preserving bacterial structure and essential nutrients is to kill OP50 with paraformaldehyde (PFA)16. PFA is a polymer of formaldehyde that can crosslink proteins within cells17 to prevent bacterial replication without destroying internal cell structures like the inner plasma membrane18. Due to this preservation of internal cellular structure, PFA-treated bacteria exhibit no growth or metabolic activity but remain an edible and nutrient-rich food source for C. elegans16. Here, a detailed protocol is provided which shows how to metabolically inactivate bacteria using paraformaldehyde.
| Method | Required Materials | Scalable? | Nutritional? | Effects on Worm? | ||||
| UV | UV-crosslinker | Limited by: | Yes | Variable effects on lifespan on NGM12, 23, 24 | ||||
| Number of plates that fit in UV-crosslinker | Variable effects on lifespan on FUdR24, 26, 27 | |||||||
| Irradiation time per plate | Decreased food preference16 | |||||||
| Ability to check every plate for growth8 | ||||||||
| Heat | >60 °C incubator | Yes | No: destroys cell wall, decreased nutritional value | Developmental arrest 13 | ||||
| Decreased food preference13 | ||||||||
| Extends lifespan on NGM31 | ||||||||
| Antibiotics | Antibiotics (kanamycin, carbenicillin, etc.) | Yes | Yes | Delays growth and development15 | ||||
| Extends lifespan in liquid media19 | ||||||||
| Extends lifespan on NGM15 | ||||||||
| PFA | 0.5% Paraformaldehyde | Yes | Yes | Small brood size decrease16 | ||||
| Small development time increase16 | ||||||||
| Decreased food preference16 | ||||||||
Table 1. Comparisons of methods to kill OP50. UV-killing, heat-killing, antibiotic-treatment, and PFA-treatment have varied effects on the nutritional status of the bacteria and the health of worms fed treated bacteria. These methods for replicatively inactivating E. coli also differ in their required materials and scalability.
Protokół
1. Bacteria inoculation
- Prepare Luria broth (LB) by dissolving 10 g of tryptone, 5 g of yeast extract, and 10 g of sodium chloride (NaCl) in 950 mL of distilled water.
- Adjust the pH of the LB to 7.0 by adding 5M sodium hydroxide (NaOH). This should only require about 0.2 mL of NaOH.
- Autoclave the pH-adjusted LB media on a liquid cycle for 45 min at 15 psi. Allow the solution to cool and store at room temperature.
- Inoculate a single colony of bacteria in 100 mL of LB in a 500 mL Erlenmeyer flask. Culture the bacteria overnight in a 37 °C shaker incubator.
- Depending on the health of the bacterial colony, the size of the flask and the speed that the shaker is set to, the time that it takes for the bacteria to grow may vary. After ~14 h, check the optical density (OD) of the bacteria at 600 nm (OD600).
- Remove the bacteria from the shaker when the OD600 is 3.0 (1 x 109 colony forming units (CFU)/mL). If the OD600 is less than 3.0, return the flask to the shaker incubator until the desired OD is reached.
- Aliquot the bacteria in 50 mL conical tubes and store at 4 °C or proceed to the next step.
2. Working with paraformaldehyde
NOTE: The concentration of paraformaldehyde (PFA) used, and the duration of exposure may vary somewhat depending on climate, location, and type of bacteria being treated. A good starting point for OP50 is exposure to 0.5% PFA for 1 h, whereas 0.25% PFA for 1 h may be sufficient for HT115.
- Prepare 32% PFA stock or use commercially purchased 32% PFA solution. Use proper personal protective equipment (PPE) when working with PFA. Wear gloves and eye protection.
- Add PFA inside a chemical fume hood with proper ventilation. Dispose of PFA containing washes and tips in proper chemical hazard containers in the fume hood.
3. Bacterial treatment with paraformaldehyde
- Once the bacteria reach an OD600 of 3.0, use a serological pipette to transfer 50 mL into a new 250 mL Erlenmeyer flask. Save the rest for a live control, a mock-treated control (see step 4), or to treat as well with PFA as needed.
- Avoid pouring from one flask to another and be cautious not to splash the sides of the new flask with bacteria. Colonies on the side of the flask may receive a lower dose of PFA.
- In the chemical hood, add 781 µL of 32% PFA to 50 mL bacteria to bring the final concentration to 0.5%. Dispose of the tip used in a solid waste chemical hazard container.
- Cover the flask with foil and return to the 37 °C shaker incubator for 1 h. After 1 h, remove the flask from the incubator and proceed to step 5.
4. Mock-treated control
- Once the bacteria reach an OD600 of 3.0, use a serological pipette to transfer 50 mL of the bacteria to a 50 mL conical tube.
- Proceed to step 5.3 to complete the washing steps similar to the PFA-treated group.
5. Washing the bacteria to remove residual PFA
- In the chemical hood, use a serological pipette to transfer the treated bacteria from the Erlenmeyer flask to a 50 mL conical tube. Using a serological pipette instead of pouring the bacteria will prevent contamination from any bacterial colonies on the edge of the flask that may have avoided direct treatment with PFA.
- Centrifuge treated bacteria at approximately 3000 x g for 20 min. Remove the supernatant by disposing of it in a liquid waste chemical hazard container in the chemical hood.
- Add 25 mL of LB and vortex to resuspend the bacterial pellet (Filling the tube fully makes it harder to resuspend the pellet). Repeat centrifugation and pellet resuspension 4x.
- Resuspend the pellet in volumes optimal for different assays. For lifespan assays, seed 60 mm plates with 200 µL of bacteria resuspended in 10 mL of LB. Resuspending the bacteria in 10 mL of LB results in a 5x concentration from the original 50 mL culture.
- Store the bacteria at 4 °C.
6. Quality check of bacterial growth
- After the final wash and resuspension, streak an LB plate (using a sterile pipette tip) with the prepared bacteria. It is good practice to streak the LB used for washing and resuspending on a separate plate as well to make sure that the LB used was not contaminated.
- Place the plates in a 37 °C incubator overnight. Check for any growth. The bacteria are considered replicatively dead when colonies do not grow on the LB plate.
7. Quality check for bacterial metabolism using a respirometer
- After the final wash and resuspension from step 5.4, confirm that the bacteria are metabolically dead using available tools such as respirometers19,20 and measuring the basal oxygen consumption rate (OCR).
- Prepare M9 solution: Dissolve 3 g of potassium phosphate monobasic (KH2PO4), 6 g of sodium phosphate dibasic (Na2HPO4), and 5 g of sodium chloride (NaCl) in 950 mL of distilled water. Autoclave on a liquid cycle for 45 min at 15 psi, then allow the solution to cool to room temperature. Add 1 mL of 1 M magnesium sulfate (MgSO4) and store at room temperature.
- Hydrate the respirometer cartridge: Add 200 µL of calibrant to all wells of a 96-well plate. Place the cartridge in the 96-well plate and incubate overnight in a 37 °C incubator.
- Assay calibration: On the following day, place the hydrated cartridge in the machine and begin calibration.
- Assay test plate setup: Using a new 96-well plate, add 160 µL of M9 and 40 µL of the prepped bacteria (1 x 109 CFU/mL) to test wells. Add 200 µL of M9 to the 4 corner wells to use as blank wells. Add 160 µL of M9 and 40 µL of LB used for washing and resuspending to use as negative controls. Add 200 µL of M9 to the rest of the wells that will not be used.
- Run the assay: Once the cartridge calibration is complete, insert the assay plate from step 7.5 into the machine for analysis. The settings include steps to mix, wait, measure and loop. The results will be shown as oxygen consumption rate (OCR). The bacteria are metabolically dead and ready to use when the OCR is zero.
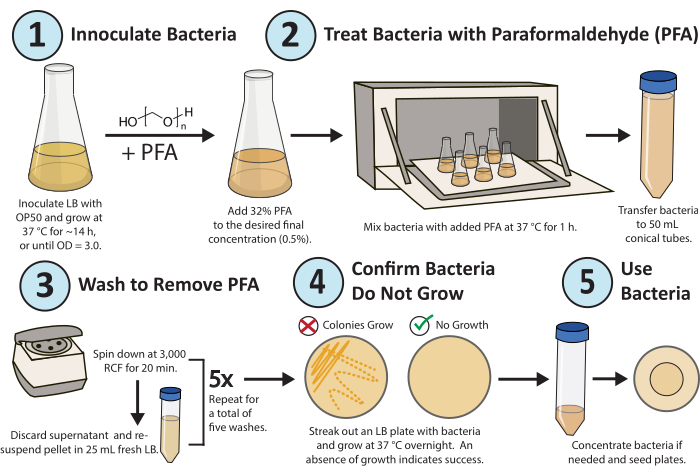
Figure 1. Workflow for paraformaldehyde treatment. A single colony of E. coli OP50 bacteria is grown overnight. PFA is added to a final concentration of 0.5%, and the PFA-treated culture is shaken for 1 h at 37 °C. Finally, the PFA is removed by washing the culture with fresh LB 5x. To confirm the treated bacteria are replicateively inactive, streak out an LB plate of the treated bacteria and grow overnight. Please click here to view a larger version of this figure.
Wyniki
A detailed workflow of the protocol is shown in Figure 1. A high-throughput method was developed and optimized to consistently inactivate bacterial replication (Figure 2A) and metabolism (Figure 2B) for metabolic and drug studies in C. elegans research using paraformaldehyde16. The goal was to determine the lowest concentration of PFA needed and the shortest amount of time required to consistently ki...
Dyskusje
Benefits of PFA-killing relative to other bacterial-killing methods
PFA-treatment is a high-throughput method to prevent bacterial metabolism while maintaining a nutritious food source for C. elegans. Killing bacteria via PFA-treatment has multiple advantages over other methods. Unlike UV-treatment, where every plate must be tested for successful killing, a single plate from a batch of PFA-treated bacteria can be tested to validate the batch16. PFA-treatment is also...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
This work was funded by NIH R21AG059117 and the Paul F. Glenn Laboratories for Biology of Aging Research at the University of Michigan. SB was funded by T32AG000114. ESK was funded by NSF DGE 1841052.
Materiały
| Name | Company | Catalog Number | Comments |
| Aluminum Foil | Staples | 2549291 | |
| Bunsen burner | VWR | 470121-700 | |
| Cell Density Meter | Denville | 80-3000-45 | |
| Centrifuge | Eppendorg | 5430 | |
| Chemical fume hood | Labcono | 975050411384RG | |
| Conincal tubes (50 mL) | Fisher | 339652 | |
| Cuvettes | Fisher | 14-955-127 | |
| E. coli OP50 | CGC | OP50 | |
| Erlenmyer flasks | Fisher | 250 mL: FB501250 500 mL: FB501500 1000 mL: FB5011000 | |
| Inoculation loop | Fisher | 22-363-605 | |
| LB Agar | Fisher | BP1425500 | |
| Liquid waste collection bottle | Thomas Scientific | 1230G50 | |
| Magnesium Sulfate (MgSO4) | Sigma | M7506 | |
| Paraformaldehyde (32%) | Electron Microscopy Sciences | 15714-S | Paraformaldehyde – methanol free solution |
| Pipettor | Eppendorf | Eppendorf Easypet 3 | |
| Plastic dishes (100 mm) | Fisher | FB0875712 | |
| Potassium Phosphate Monobasic (KH2PO4) | Fisher | P2853 | |
| Seahorse XF Calibrant | Agilent | 100840-000 | |
| Seahorse XFe96 Extracellular Flux Assay Kit and Cell Culture Microplate | Agilent | 101085-004 | |
| Serological pipettes (50 mL) | Genesee Scientific | 12-107 | |
| Shaker incubator | Thermo | 11 676 083 | |
| Sodium Chloride (NaCl) | Fisher | S640-3 | |
| Sodium Hydroxide (NaOH) | Fisher | S318500 | |
| Sodium Phosphate Dibasic Anhydrous (Na2HPO4) | Sigma | S374-500 | |
| Solid waste collection bucket | M&M Industries | 5.0 Gallon M1 Traditional Pail | |
| Tryptone | Genesee Scientific | 20-251 | |
| Vortex | Thermo | 11676331 | |
| Weighing balance | C Goldenwall | HZ10K6B | |
| Yeast Extract | Genesee Scientific | 20-255 |
Odniesienia
- Riddle, D. L., Blumenthal, T., Meyer, B. J., Priess, J. R. C. . Elegans II. 33, (1997).
- Corsi, A. K., Wightman, B., Chalfie, M. A transparent window into biology: A primer on caenorhabditis elegans. WormBook. , 1-31 (2015).
- Kaletta, T., Hengartner, M. O. Finding function in novel targets: C. elegans as a model organism. Nature reviews. Drug discovery. 5 (5), 387-398 (2006).
- Meneely, P. M., Dahlberg, C. L., Rose, J. K. Working with worms: Caenorhabditis elegans as a model organism. Current Protocols Essential Laboratory Techniques. 19 (1), (2019).
- Zhang, S., Li, F., Zhou, T., Wang, G., Li, Z. Caenorhabditis elegans as a useful model for studying aging mutations. Frontiers in Endocrinology. 11, 554994 (2020).
- Feng, M., Gao, B., Garcia, L. R., Sun, Q. Microbiota-derived metabolites in regulating the development and physiology of Caenorhabditis elegans. Frontiers in Microbiology. 14, 1035582 (2023).
- McIntyre, G., Wright, J., Wong, H. T., Lamendella, R., Chan, J. Effects of FUdR on gene expression in the C. elegans bacterial diet OP50. BMC Research Notes. 14 (1), 207 (2021).
- Cabreiro, F., et al. Metformin retards aging in c. Elegans by altering microbial folate and methionine metabolism. Cell. 153 (1), 228-239 (2013).
- Worthy, S. E., et al. Identification of attractive odorants released by preferred bacterial food found in the natural habitats of c. Elegans. PLoS One. 13 (7), e0201158 (2018).
- Chen, Y. C., Seyedsayamdost, M. R., Ringstad, N. A microbial metabolite synergizes with endogenous serotonin to trigger C. elegans reproductive behavior. Proceedings of the National Academy of Sciences of the United States of America. 117 (48), 30589-30598 (2020).
- Kim, D. H., Flavell, S. W. Host-microbe interactions and the behavior of Caenorhabditis elegans. Journal of Neurogenetics. 34 (3-4), 500-509 (2020).
- Gems, D., Riddle, D. L. Genetic, behavioral, and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 154 (4), 1597-1610 (2000).
- Qi, B., Kniazeva, M., Han, M. A vitamin-b2-sensing mechanism that regulates gut protease activity to impact animal's food behavior and growth. eLife. 6, e26243 (2017).
- Garigan, D., et al. Genetic analysis of tissue aging in caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics. 161 (3), 1101-1112 (2002).
- Virk, B., et al. Folate acts in E. coli to accelerate C. elegans aging independently of bacterial biosynthesis. Cell Reports. 14 (7), 1611-1620 (2016).
- Beydoun, S., et al. An alternative food source for metabolism and longevity studies in Caenorhabditis elegans. Communications Biology. 4 (1), 258 (2021).
- Thavarajah, R., Mudimbaimannar, V. K., Elizabeth, J., Rao, U. K., Ranganathan, K. Chemical and physical basics of routine formaldehyde fixation. Journal of Oral and Maxillofacial Pathology. 16 (3), 400-405 (2012).
- Felix, H. Permeabilized and immobilized cells. Methods in Enzymology. 137, 637-641 (1988).
- Lobritz, M. A., et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proceedings of the National Academy of Sciences of the United States of America. 112 (27), 8173-8180 (2015).
- Nadanaciva, S., et al. Assessment of drug-induced mitochondrial dysfunction via altered cellular respiration and acidification measured in a 96-well platform. Journal of Bioenergetics and Biomembranes. 44 (4), 421-437 (2012).
- Shtonda, B. B., Avery, L. Dietary choice behavior in Caenorhabditis elegans. The Journal of Experimental biology. 209 (Pt 1), 89-102 (2006).
- MacNeil, L. T., Watson, E., Arda, H. E., Zhu, L. J., Walhout, A. J. Diet-induced developmental acceleration independent of tor and insulin in C. elegans. Cell. 153 (1), 240-252 (2013).
- Kumar, S., et al. Lifespan extension in C. elegans caused by bacterial colonization of the intestine and subsequent activation of an innate immune response. Developmental Cell. 49 (1), 100-117 (2019).
- Nakagawa, H., et al. Effects and mechanisms of prolongevity induced by Lactobacillus gasseri sbt2055 in Caenorhabditis elegans. Aging Cell. 15 (2), 227-236 (2016).
- Kaeberlein, T. L., et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 5 (6), 487-494 (2006).
- Beaudoin-Chabot, C., et al. The unfolded protein response reverses the effects of glucose on lifespan in chemically-sterilized C. elegans. Nature Communication. 13 (1), 5889 (2022).
- Komura, T., Takemoto, A., Kosaka, H., Suzuki, T., Nishikawa, Y. Prolonged lifespan, improved perception, and enhanced host defense of Caenorhabditis elegans by Lactococcus cremoris subsp. cremoris.Microbiology Spectrum. 10 (3), e0045421 (2022).
- Ye, X., Linton, J. M., Schork, N. J., Buck, L. B., Petrascheck, M. A pharmacological network for lifespan extension in Caenorhabditis elegans. Aging Cell. 13 (2), 206-215 (2014).
- Hastings, J., et al. Wormjam: A consensus C. elegans metabolic reconstruction and metabolomics community and workshop series. Worm. 6 (2), e1373939 (2017).
- O'Donnell, M. P., Fox, B. W., Chao, P. H., Schroeder, F. C., Sengupta, P. A neurotransmitter produced by gut bacteria modulates host sensory behaviour. Nature. 583 (7816), 415-420 (2020).
- Stuhr, N. L., Curran, S. P. Bacterial diets differentially alter lifespan and healthspan trajectories in C. elegans. Communications Biology. 3 (1), 653 (2020).
- Dirksen, P., et al. Cembio - the Caenorhabditis elegans microbiome resource. G3 (Bethesda). 10 (9), 3025-3039 (2020).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone