Culturas puras e semeadura por esgotamento: isolamento de colônias bacterianas únicas de uma amostra mista
Visão Geral
Fonte: Tilde Andersson1, Rolf Lood1
1 Departamento de Ciências Clínicas Lund, Divisão de Medicina de Infecção, Centro Biomédico, Universidade de Lund, 221 00 Lund, Suécia
Aparentemente impossível de determinar, a biodiversidade microbiana é verdadeiramente surpreendente com uma estimativa de um trilhão de espécies coexistindo (1,2). Embora climas particularmente severos, como o ambiente ácido do estômago humano (3) ou os lagos subglaciais da Antártida (4), possam ser dominados por uma espécie específica, as bactérias são tipicamente encontradas em culturas mistas. Como cada cepa pode influenciar o crescimento de outra (5), a capacidade de separar e cultivar colônias "puras" (consistindo apenas de um tipo) tornou-se essencial em ambientes clínicos e acadêmicos. Culturas puras permitem mais exames genéticos (6) e proteômicos (7), análise da pureza amostral e, talvez mais notável, a identificação e caracterização de agentes infecciosos a partir de amostras clínicas.
As bactérias têm uma ampla gama de requisitos de crescimento e existem inúmeros tipos de mídia de nutrientes projetadas para sustentar tanto as espécies não exigentes quanto as meticulosas (8). A mídia de crescimento pode ser preparada na forma líquida (como um caldo) ou em uma forma sólida tipicamente baseada em ágar (um agente de gelling derivado de algas vermelhas). Considerando que a inoculação direta no caldo traz o risco de gerar uma população bacteriana geneticamente diversa ou mesmo mista, chapeamento e re-streaking cria uma cultura mais pura onde cada célula tem uma composição genética altamente semelhante. A técnica da placa de raia baseia-se na diluição progressiva de uma amostra(Figura 1),com o objetivo de separar células individuais umas das outras. Qualquer célula viável (doravante referida como uma unidade formadora de colônias, CFU) sustentada pela mídia e ambiente designado pode posteriormente encontrar uma colônia isolada de células filhas através de fissão binária. Apesar das rápidas taxas de mutação dentro das comunidades bacterianas, esse grupo celular é geralmente considerado clonal. A colheita e re-streaking dessa população, consequentemente, garante que o trabalho subsequente envolva apenas um único tipo de bactéria.
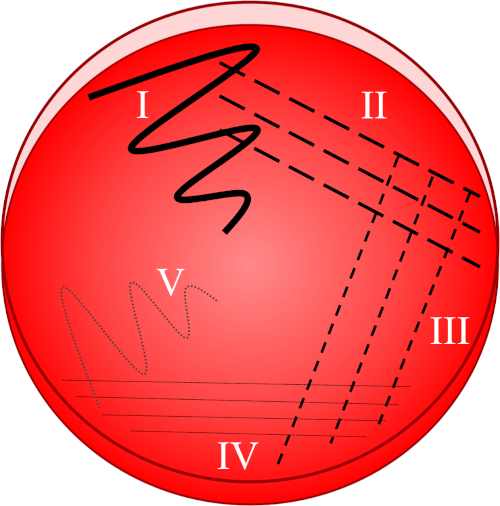
Figura 1: Uma placa de raia é baseada na diluição progressiva da amostra original. I) O inóculo é inicialmente disperso usando um movimento em zig-zag, criando uma área com uma população bacteriana relativamente densa. II-IV) As listras são extraídas da área anterior, usando um laço de inoculação estéril cada vez, até que o quarto quadrante seja atingido. V) Um movimento final em zigue-zague direcionado para o meio da placa forma uma região onde o inóculo foi marcadamente diluído, permitindo que as colônias apareçam separadas umas das outras.
A técnica da placa de raia também pode ser combinada com o uso de mídias seletivas e/ou diferenciais. Um meio seletivo inibirá o crescimento de certos organismos ( porexemplo, através da adição de antibióticos), enquanto um meio diferencial só ajudará a distinguir um do outro (por exemplo, através da hemolise em placas de ágar sanguíneo).
Por trás de todo o trabalho em microbiologia está o uso de técnicas assépticas (estéreis). Toda cultura bacteriana deve ser considerada potencialmente patogênica, pois há risco de crescimento não intencional de cepas traiçoeiras, formação de aerossol e contaminação de equipamentos/pessoal. Para minimizar esses riscos, todas as mídias, plásticos, metal e vidro são tipicamente esterilizados através de autoclaving antes e depois do uso, submetendo-os a vapor saturado de alta pressão em torno de 121°C que efetivamente elimina quaisquer células persistentes. O espaço de trabalho é geralmente desinfetado usando etanol antes e depois do uso. Jaleco e luvas são sempre usados durante o trabalho com agentes infecciosos.
Procedimento
1. Configurar
- Todos os micróbios devem ser tratados como se fossem perigosos. Use sempre um jaleco e luvas, amarre os cabelos longos e certifique-se de que todas as feridas estejam particularmente bem protegidas.
- Prepare o espaço de trabalho esterilizando-o usando 70% de etanol.
- Certifique-se de que as placas de ágar, a solução de amostra(s) e uma caixa de laços de inoculação de plástico pré-esterilizados ou uma alça metálica mais uma chama bunsen, estão próximas. Alças de p
Resultados
A placa de listras inicial pode conter colônias originárias de células com diferentes maquiagens genéticas ou (dependendo da pureza da amostra) de diferentes espécies bacterianas(Figura 2A).
Através do isolamento subsequente de uma única colônia, onde todas as unidades são derivadas de uma célula-mãe comum, o segundo procedimento de estria gera uma população bacteriana ...
Aplicação e Resumo
A capacidade de obter e cultivar uma colônia bacteriana pura é essencial, tanto em ambientes clínicos quanto acadêmicos. O revestimento de raia permite o isolamento de uma população celular relativamente clonal, originária de uma UFC compartilhada, que pode ser de particular interesse durante o diagnóstico ou para caracterização adicional do isolado. Uma amostra é listrada em um meio de nutrientes à base de ágar adequado e incubada até que as colônias se tornem visíveis. Uma colônia isolada é posteriorm...
Referências
- The Human Microbiome Project C. Structure, Function and Diversity of the Healthy Human Microbiome. Nature. 486:207-214. (2012)
- Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. Proceedings of the National Academy of Sciences. 113 (21) 5970-5975 (2016)
- Skouloubris S, Thiberge JM, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infection and Immunity. 66:4517-21. (1998)
- Mikucki JA, Auken E, Tulaczyk S, Virginia RA, Schamper C, Sørensen KI, Doran PT, Dugan H, Foley N. Deep groundwater and potential subsurface habitats beneath an Antarctic dry valley. Nature Communications. 6:6831. (2015)
- Mullineaux-Sanders C, Suez J, Elinav E, Frankel G. Sieving through gut models of colonization resistance. Nature Microbiology. 3:132-140. (2018)
- Fournier PE, Drancourt M, Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infectious Diseases. 7:711-23 (2007)
- Yao Z, Li W, Lin Y, Wu Q, Yu F, Lin W, Lin X. Proteomic Analysis Reveals That Metabolic Flows Affect the Susceptibility of Aeromonas hydrophila to Antibiotics. Scientific Reports. 6:39413 (2016)
- Medina D, Walke JB, Gajewski Z, Becker MH, Swartwout MC, Belden LK. Culture Media and Individual Hosts Affect the Recovery of Culturable Bacterial Diversity from Amphibian Skin. Frontiers in Microbiology. 8:1574 (2017)
Pular para...
Vídeos desta coleção:

Now Playing
Culturas puras e semeadura por esgotamento: isolamento de colônias bacterianas únicas de uma amostra mista
Microbiology
166.2K Visualizações

Criando uma coluna de Winogradsky: um método para enriquecer as espécies microbianas em uma amostra de sedimento
Microbiology
129.5K Visualizações

Diluições em série e plaqueamento: enumeração microbiana
Microbiology
316.3K Visualizações

Culturas de enriquecimento: cultivo de micróbios aeróbicos e anaeróbicos em meios seletivos e diferenciais
Microbiology
132.1K Visualizações

Sequenciamento de rRNA 16S: uma técnica baseada em PCR para identificar espécies bacterianas
Microbiology
189.1K Visualizações

Curvas de crescimento: gerando curvas de crescimento usando unidades formadoras de colônias e medições de densidade óptica
Microbiology
296.5K Visualizações

Teste de sucetibilidade a antibióticos: testes de epsilômetro para determinar valores de MIC de dois antibióticos e avaliar a sinergia de antibióticos
Microbiology
93.8K Visualizações

Microscopia e Coloração: Coloração de Gram, Cápsula e Endósporo
Microbiology
363.5K Visualizações

Ensaio de placa: um método para determinar o título viral como unidades formadoras de placa (PFU)
Microbiology
186.3K Visualizações

Transformação de células de E. coli usando um protocolo adaptado de cloreto de cálcio
Microbiology
86.9K Visualizações

Conjugação: um método para transferir a resistência à ampicilina da E. coli doadora para a receptora
Microbiology
38.3K Visualizações

Transdução fágica: um método para transferir a resistência à ampicilina da E. coli doadora para a receptora
Microbiology
29.1K Visualizações
Copyright © 2025 MyJoVE Corporation. Todos os direitos reservados