A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Modified Yeast-Two-Hybrid System to Identify Proteins Interacting with the Growth Factor Progranulin
In This Article
Summary
We have modified the conventional yeast two-hybrid screening, an effective genetic tool in identifying protein interaction. This modification markedly shortens the process, reduces the workload, and most importantly, reduces the number of false positives. In addition, this approach is reproducible and reliable.
Abstract
Progranulin (PGRN), also known as granulin epithelin precursor (GEP), is a 593-amino-acid autocrine growth factor. PGRN is known to play a critical role in a variety of physiologic and disease processes, including early embryogenesis, wound healing 1, inflammation 2, 3, and host defense 4. PGRN also functions as a neurotrophic factor 5, and mutations in the PGRN gene resulting in partial loss of the PGRN protein cause frontotemporal dementia 6, 7. Our recent studies have led to the isolation of PGRN as an important regulator of cartilage development and degradation 8-11. Although PGRN, discovered nearly two decades ago, plays crucial roles in multiple physiological and pathological conditions, efforts to exploit the actions of PGRN and understand the mechanisms involved have been significantly hampered by our inability to identify its binding receptor(s). To address this issue, we developed a modified yeast two-hybrid (MY2H) approach based on the most commonly used GAL4 based 2-hybrid system. Compared with the conventional yeast two-hybrid screen, MY2H dramatically shortens the screen process and reduces the number of false positive clones. In addition, this approach is reproducible and reliable, and we have successfully employed this system in isolating the binding proteins of various baits, including ion channel 12, extracellular matrix protein 10, 13, and growth factor14. In this paper, we describe this MY2H experimental procedure in detail using PGRN as an example that led to the identification of TNFR2 as the first known PGRN-associated receptor 14, 15.
Protocol
1. Background information
The yeast two-hybrid system is a powerful genetic technique used to discover protein-protein interactions 16, 17. Several kinds of 2-hybrid systems, such as lexA-based systems, the Sos Recruitment System, and bacteria- or mammalian cell-based 2-hybrids, are commercial available, this paper specifically focuses on the modifications of the most commonly used GAL4 based yeast 2-hybrid system. Briefly, the method is based on the properties of the yeast GAL4 protein that consists of separable domains responsible for DNA-binding and transcriptional activation. The bait protein is expressed as a fusion to the GAL4 DNA-binding domain (DNA-BD), while the prey proteins are expressed as fusions to the GAL4 activation domain (AD). Interaction between bait and prey fusion proteins leads to the transcriptional activations of GAL4-binding sites containing reporter genes that are integrated into the yeast genome. The principle of Y2H is illustrated in Fig. 1 and the experimental procedure is summarized in Fig. 2.
2. Required materials and solutions
- YPD Growth Medium (a blend of peptone, yeast extract, and dextrose in optimal proportions for growing most Saccharomyces cerevisiae strains).
- Minimal SD Bases (Minimal synthetic defined (SD) bases include a yeast nitrogen base, ammonium sulfate, and a carbon source, dextrose. Dropout (DO) supplements can be added to the Minimal SD Base to make a synthetic, defined medium lacking the specified nutrients).
- Leu/-Trp Dropout (DO) supplement (containing every essential amino acid except for leucine and tryptophan)
- -His/-Leu/-Trp/-Ura Dropout (DO) Supplement (containing every essential amino acid except for leucine, tryptophan, histidine, and uracil)
- Luria broth (LB) (Tryptone 10 g/L, Yeast extract 5 g/L, NaCl 5 g/L)
- X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) solution: prepared as a 20 mg/ml stock solution in N, N-Dimethylformamide
- 10X TE (100 mM Tris-HCl (pH 7.5), 10 mM EDTA, autoclaved)
- Sonicated Herring or Salmon Sperm DNA, boiled (10 mg/ml)
- 10X LiAc (1 M lithium acetate, autoclaved)
- 50% PEG-3350 solution, filter-sterilized
- 3-Amino-1, 2, 4-Triazole (3AT)
- Kanamycin
- Ampicillin
- ELECTROMAX DH5α cells
- Z buffer: 16.1 g Na2HPO4 7H2O (or 8.52 g anhydrous), 5.5 g NaH2PO4 H2O (or 4.8 ganhydrous), 0.75 g KCl, 0.246 g MgSO4 7H2O (or 0.12 g anhydrous), dissolved in 1 L autoclaved, distilled water and adjusted to pH 7.0.
3. Bait generation (pDBLeu-PGRN)
A cDNA fragment encoding PGRN lacking signal peptide (a.a.21-588) was directionally cloned into the Sal I-Not I sites of the pDBLeu vector (the ProQuest two-hybrid system, Invitrogen), keeping the same translation reading frame as the GAL4 DNA Binding Domain to generate pDBLeu-PGRN.
- Amplify the cDNA fragment of PGRN described above by PCR using oligonucleotide primers designed to contain restriction sites (Sal I at the 5′end and Not I at the 3′end) to allow in-frame fusion.
- Gel purify the PCR product, digest with restriction endonucleases Sal I and Not I.
Prepare the DB vector pDBLeu by double restriction digestion with the same restriction endonucleases. - Ligate the restriction PGRN fragment into the linearized pDBLeu vector and transform into DH5α with selection for LB+kanamycin at 25 μg/ml.
- Verify the correction of the construct by DNA sequencing.
4. Small scale transformation of bait plasmid
- Inoculate 5 ml of YPD with a colony of Mav203 (Invitrogen), shake overnight at 30°C.
- Dilute the overnight culture in 50 ml of YPD. Grow an additional 2-4 hours.
- Pellet the cells at 3000 rpm for 5 min at RT. Resuspend the pellet in 40 ml of autoclaved, distilled water.
- Re-pellet the cells. Resuspend in 2 ml of solution I, incubate at RT 10 min.
Solution I: 0.5 ml 10xLiAc, 0.5 ml 10xTE, 4 ml H2O - Dispense the DNA to tubes: 2-3 μl of pDBLeu-PGRN (0.25μg/μl) and 10 μl of denatured, sheared salmon sperm DNA (10μg/μl). Add 100μl yeast cells, mix well.
- Add 700μl solution II, mix well. Incubate at 30°C for 30 min. Solution II: 0.2 ml 10xLiAc, 0.2 ml 10xTE, 1.6 ml 50% PEG3350.
- Heat shock at 42°C for 15 min.
- Pellet for 2 min. Discard the supernatant. Resuspend the pellet in 200 μl autoclaved, distilled water.
- Plate the suspension onto SD-Leu plates with serial dilutions. Incubate the plate at 30°C for 2-3 days.
5. Bait validation
Before performing yeast two-hybrid screen, test pDBLeu-PGRN for self-activation and determine the basal expression levels of the HIS3 reporter gene. This test determines whether baits activate transcription and whether the self-activation can be neutralized by inhibitors. 3-Amino-1, 2, 4-triazole (3-AT) is a competitive inhibitor of the HIS3-gene product and can be used to titrate the minimum level of HIS3 expression required for growth on histidine-deficient media.
- Transform the pDBLeu-PGRN into yeast Mav203 strain, plate the transformation onto SD-Leu plates, and incubate for 48-72 h at 30°C.
- Patch colonies from each transformation using autoclaved toothpicks onto SD-Leu-His plates containing 3-AT at concentrations of 10 mM, 25 mM, 50 mM, 75 mM, and 100 mM. 3-AT is a competitive inhibitor of the HIS3 enzyme and only baits that exhibit self-activation will be able to grow in the presence of the 3-AT.
- Bait strains that grow on plates containing 100 mM 3-AT are not suitable for use in the two-hybrid screen. For baits that can be used, use the lowest 3-AT concentration that inhibits cell growth. In many cases, 25 mM of 3-AT is used for screening.
6. cDNA library screening
The pDBLeu-PGRN plasmid is introduced into MaV203 using a small-scale transformation as described above. To introduce pPC86-library (Invitrogen) into MaV203 (pDBLeu-PGRN), the procedure described below typically gives ~4 x 104 colonies with 0.5 μg of plasmid library DNA. Hence, 2.5 x 106 yeast transformants will require ~30.0 μg pPC86-cDNA library plasmid DNA, 25 transformations, and fifty 10-cm plates (SD-Leu-Trp-His-Ura+3AT).
- Prepare the appropriate number of 10-cm SD-Leu-Trp-His-Ura+3AT plates (50 for the procedure below). Also prepare at least four 10-cm SD-Leu-Trp plates for estimating the number of transformants.
- Transform the library into the MaV203 containing pDBLeu-PGRN according to the procedure described below.
- Suspend several isolated colonies of MaV203 (pDBLeu-PGRN) in ~100μl autoclaved, distilled water and spread them onto a 10-cm SD-Leu plate. Repeat procedure for a second SD-Leu plate. Incubate both plates for 18-24 h at 30°C.
- Scrape and completely suspend the cells in 10 ml autoclaved, distilled water. Add a sufficient volume of cell suspension to 500 ml of liquid YPD medium in a flask to give an OD600 of ~0.1.
- Verify that the OD is ~0.1 after inoculation.
- Shake at 30°C until the OD600 reaches ~0.4.
- Prepare fresh:
- 110 ml 1X TE/LiAc by combining 11 ml 10X TE, 11 ml 10X LiAc, and 88 ml autoclaved water.
- 16 ml PEG/LiAc by combining 1.6 ml 10X TE, 1.6 ml 10X LiAc, and 12.8 ml 50% PEG-3350.
- Pellet the cells at 3000g for 5 min at room temperature.
- Discard the supernatants and gently resuspend the pellet by pipetting up and down in 100 ml autoclaved, distilled water at room temperature.
- Centrifuge at 3000 g for 5 min at room temperature.
- Discard the supernatant of the centrifuged cells and resuspend the cell pellet in 50 ml 1X TE/LiAc solution.
- Centrifuge at 3000 g for 5 min at room temperature.
- Remove the supernatants and resuspend the pellet in a final volume of 2.5 ml 1X TE/LiAc solution.
- Perform 25 transformations. Combine 2.5 ml cells, 125 μl (10μg/μl) denatured, sheared salmon sperm DNA, and 30 μg cDNA library. Mix gently by pipetting up and down. Add 15 ml PEG/LiAc solution and mix gently. Aliquot into 25 autoclaved 1.5-ml microcentrifuge tubes of 700 μl each.
- Incubate for 30 min in a 30°C water bath.
- Heat shock for 15 min in a 42°C water bath.
- Centrifuge at 6,000 g for 1 min at room temperature. Carefully remove the supernatant. Gently resuspend each pellet in 400 μl autoclaved, distilled water by pipetting up and down.
- Plate the 200 μl transformation mixture from each transformation onto single 10-cm SD-Leu-Trp-His-Ura+3AT (3AT concentration: 25 mM) plates using a spread bar, so 25 autoclaved 1.5-ml microcentrifuge tubes can make 50 plates (SD-Leu-Trp-His-Ura +3AT).
- Incubate plates for 5-10 days at 30°C.
- To estimate the transformation efficiency of the reaction, serial dilutions of one reaction (1:40; 1:400 and 1:4000) are plated on SD-Leu-Trp plate. After incubating for 3 days at 30°C, the number of colonies is counted and the total number of transformants calculated.
7. X-gal assay
- Transfer colonies to YPD plate; incubate at 30°C overnight.
- Place rounded Nitrocellulose membrane on the YPD plate; make sure there are no bubbles between the membrane and the YPD plate. After 1-2 min, make sure all colonies are transferred to membranes (nitrocellulose paper).
- Place the membrane colony side up in boat made with aluminum foil paper.
- Put the membrane into liquid nitrogen, wait twenty seconds, and submerge into liquid nitrogen for a couple of minutes.
- Take out membrane to thaw.
- Meanwhile mix the following reagent:
1.5ml Z buffer
20μl X-gal (20 mg/ml) - Drop Z buffer/X-gal in a petri dish, place whatman paper on top of that.
- Carefully place nitrocellulose paper on top of whatman paper.
- Incubate at 37° until blue colonies appear.
8. Retransformation assay
Prey fusion proteins (AD-Y) isolated from library screening should retain the interaction with bait fusion protein (DB-X) to induce the report genes, and the retransformation of prey clones and bait construct into yeast can further eliminate false positives and facilitate additional analysis.
- Isolate plasmid DNA from yeast strains potentially containing interacting proteins.
- Transform DNA into E. coli DH5α cells, plate the transformations on LB+100 μg/ml ampicillin plates to selectively isolate the pPC86-cDNA library, and incubate overnight at 37°C.
- Pick several colonies from the plate to put into LB+100 μg/ml ampicillin broth.
- Prepare miniprep DNA and examine by double restriction analysis with Sal I and Not I.
- Co-transform the prey plasmid and bait plasmid into MaV203, plate the transformation mixtures on SC-Leu-Trp plates, and incubate for 2-3 days at 30°C.
- Pick three different colonies from the SC-Leu-Trp plate, perform X-gal Assay.
9. Sequencing and bioinformatics analysis
Sequence the plasmid DNA that was isolated from likely true positive clones, compare these sequences to those in the GenBank using the BLAST program, and identify those two-hybrid clones that correspond to known genes. Sequencing data showed that two of the 12 positives obtained above were cell surface TNFR2 (TNFRSF1B/CD120b; Accession #NM_130426). In addition, the interaction between PGRN and TNFR was verified using various protein-protein interaction assays, including in vitro Solid-phase binding assay, Co-immunoprecipitation, Surface plasmon resonance analysis, and Flow cytometry assay 14.
10. Representative Results
The flow chart of the screening is outlined in Fig. 3. Typically 50-100 positive clone candidates will be obtained at this step. We initially isolated 54 positive clone candidates among 2.5 million transformants screened with the PGRN bait. Positive clone candidates were then verified by performing X-gal assay. Typically, approximately 50% false positive clones are removed via X-gal assay. We obtained 23 positive clone candidates at this step for the PGRN bait (Fig. 4). Retransformation of the prey clones and bait construct into yeast further eliminate false positives and clones that still activate the reporter genes likely represent the true positives. Typically, approximately 50% positive clone candidates will be removed. We finally isolated 12 positive clones that interact with PGRN in yeast.
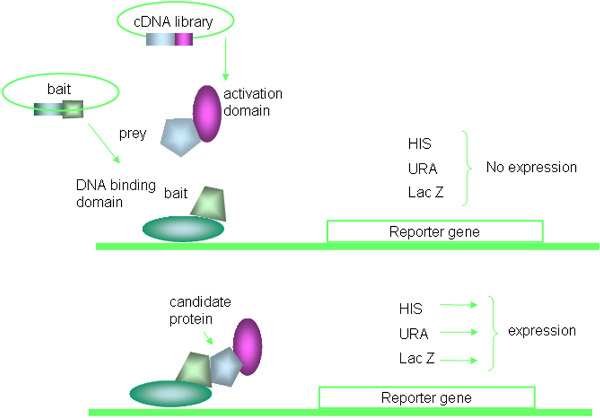
Figure 1. Principle of yeast two-hybrid system

Figure 2. Pipeline of identifying protein binding partners using yeast two-hybrid system

Figure 3. Flow chart of screening yeast two-hybrid library. Click here to view a full-sized version of this image.
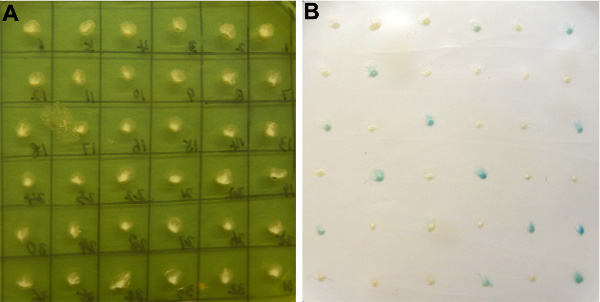
Figure 4. Beta-Galactosidase assay of positive clone candidates. A, Positive clones obtained from library screen were transferred to YPD plate and incubated at 30°C overnight; B, All colonies on YPD plate were transferred to nitrocellulose membranes and beta-galactosidase assay performed.
Access restricted. Please log in or start a trial to view this content.
Discussion
Yeast two-hybrid screening has proven to be an effective tool in identifying protein interaction 16, 17. Compared with other approaches for identifying protein-binding partners, such as biochemical co-purification and protein chips, yeast two-hybrid system is a sensitive genetic approach that can be used for screening very high numbers of coding sequences in a relatively simple experiment; in addition, it detects the in vivo interaction and does not need complicated protein purification. Of course yeast two-hy...
Access restricted. Please log in or start a trial to view this content.
Disclosures
No conflicts of interest declared.
Acknowledgements
This work was funded by NIH research grants K01AR053210, R01AR061484 and a grant from National Psoriasis Foundation.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| ProQuest two-hybrid system | Invitrogen | 10835 | |
| YPD Growth Medium | Clontech Laboratories | 630409 | |
| YPD Agar Medium | Clontech Laboratories | 630410 | |
| Minimal SD agar Base | Clontech Laboratories | 630412 | |
| -Leu DO Supplement | Clontech Laboratories | 630414 | |
| -Leu/-Trp DO Supplement | Clontech Laboratories | 630417 | |
| -His/-Leu/-Trp/-Ura DO Supplement | Clontech Laboratories | 630425 | |
| 3-Amino-1,2,4-Triazole (3AT) | Sigma-Aldrich | A8056 | |
| Luria broth (LB) | Sigma-Aldrich | L3022 | |
| X-Gal | Invitrogen | 15520-034 | |
| Sonicated Salmon Sperm DNA | Stratagene, Agilent Technologies | 201190 | |
| Ampicillin | AMRESCO | 0339 | |
| Kanamycin Sulfate | Invitrogen | 11815-024 | |
| Subcloning Efficiency™ DH5α™ Competent Cells | Invitrogen | 18265-017 | |
| Trizma® base | Sigma-Aldrich | T6066 | |
| Lithium acetate | Sigma-Aldrich | L4158 | |
| Ethylenediaminetetraacetic acid | Sigma-Aldrich | ED | |
| Nitrocellulose Membrane | Bio-Rad | 162-0115 | |
| 10-cm petri dish | ITI Scientific | CT-903 | |
| Incubator (30 °C) | ATR (Ecotron) |
References
- He, Z. Progranulin is a mediator of the wound response. Nat. Med. 9, 225-229 (2003).
- Kessenbrock, K. Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J. Clin. Invest. 118, 2438-2447 (2008).
- Zhu, J. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell. 111, 867-878 (2002).
- Yin, F. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J. Exp. Med. 207, 117-128 (2010).
- Van Damme, P. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J. Cell. Biol. 181, 37-41 (2008).
- Baker, M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 442, 916-919 (2006).
- Cruts, M. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 442, 920-924 (2006).
- Guo, F. Granulin-epithelin precursor binds directly to ADAMTS-7 and ADAMTS-12 and inhibits their degradation of cartilage oligomeric matrix protein. Arthritis Rheum. 62, 2023-2036 (2010).
- Feng, J. Q. Granulin epithelin precursor: a bone morphogenic protein 2-inducible growth factor that activates Erk1/2 signaling and JunB transcription factor in chondrogenesis. FASEB. J. 24, 1879-1892 (2010).
- Xu, K. Cartilage oligomeric matrix protein associates with granulin-epithelin precursor (GEP) and potentiates GEP-stimulated chondrocyte proliferation. J. Biol. Chem. 282, 11347-11355 (2007).
- Liu, C. J. The role of ADAMTS-7 and ADAMTS-12 in the pathogenesis of arthritis. Nat. Clin. Pract. Rheumatol. 5, 38-45 (2009).
- Liu, C., Dib-Hajj, S. D., Waxman, S. G. Waxman, S.G. Fibroblast growth factor homologous factor 1B binds to the C terminus of the tetrodotoxin-resistant sodium channel rNav1.9a (NaN). J. Biol. Chem. 276, 18925-18933 (2001).
- Liu, C. J. ADAMTS-7: a metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB. J. 20, 988-990 (2006).
- Tang, W. The Growth Factor Progranulin Binds to TNF Receptors and Is Therapeutic Against Inflammatory Arthritis in Mice. Science. 332, 478-484 (2011).
- Wu, H., Siegel, R. M. Progranulin Resolves Inflammation. Science. 332, 427-428 (2011).
- Hollenberg, S. M. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15, 3813-3822 (1995).
- Vojtek, A. B., Hollenberg, S. M., Cooper, J. A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 74, 205-214 (1993).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved