A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Two- and Three-Dimensional Live Cell Imaging of DNA Damage Response Proteins
In This Article
Summary
This protocol describes a method for visualizing a DNA double-strand break signaling protein activated in response to DNA damage as well as its localization during mitosis.
Abstract
Double-strand breaks (DSBs) are the most deleterious DNA lesions a cell can encounter. If left unrepaired, DSBs harbor great potential to generate mutations and chromosomal aberrations1. To prevent this trauma from catalyzing genomic instability, it is crucial for cells to detect DSBs, activate the DNA damage response (DDR), and repair the DNA. When stimulated, the DDR works to preserve genomic integrity by triggering cell cycle arrest to allow for repair to take place or force the cell to undergo apoptosis. The predominant mechanisms of DSB repair occur through nonhomologous end-joining (NHEJ) and homologous recombination repair (HRR) (reviewed in2). There are many proteins whose activities must be precisely orchestrated for the DDR to function properly. Herein, we describe a method for 2- and 3-dimensional (D) visualization of one of these proteins, 53BP1.
The p53-binding protein 1 (53BP1) localizes to areas of DSBs by binding to modified histones3,4, forming foci within 5-15 minutes5. The histone modifications and recruitment of 53BP1 and other DDR proteins to DSB sites are believed to facilitate the structural rearrangement of chromatin around areas of damage and contribute to DNA repair6. Beyond direct participation in repair, additional roles have been described for 53BP1 in the DDR, such as regulating an intra-S checkpoint, a G2/M checkpoint, and activating downstream DDR proteins7-9. Recently, it was discovered that 53BP1 does not form foci in response to DNA damage induced during mitosis, instead waiting for cells to enter G1 before localizing to the vicinity of DSBs6. DDR proteins such as 53BP1 have been found to associate with mitotic structures (such as kinetochores) during the progression through mitosis10.
In this protocol we describe the use of 2- and 3-D live cell imaging to visualize the formation of 53BP1 foci in response to the DNA damaging agent camptothecin (CPT), as well as 53BP1's behavior during mitosis. Camptothecin is a topoisomerase I inhibitor that primarily causes DSBs during DNA replication. To accomplish this, we used a previously described 53BP1-mCherry fluorescent fusion protein construct consisting of a 53BP1 protein domain able to bind DSBs11. In addition, we used a histone H2B-GFP fluorescent fusion protein construct able to monitor chromatin dynamics throughout the cell cycle but in particular during mitosis12. Live cell imaging in multiple dimensions is an excellent tool to deepen our understanding of the function of DDR proteins in eukaryotic cells.
Protocol
A. Cell Preparation
- Normal human primary fibroblasts (GM02270) were obtained from Coriell Cell Repository, Camden, New Jersey, and immortalized with hTERT 6. Cells were grown and expanded in CellStar 6-cm dishes in media (4 ml) consisting of MEM supplemented with 20% fetal bovine serum (GIBCO), non-essential/essential amino acids, vitamins, sodium pyruvate, and penicillin/streptomycin (HyClone).
- Human embryonic kidney 293 (HEK293) cells were obtained from American Type Culture Collection and grown and expanded in CellStar 6-cm dishes. The cells were kept in media (4 ml) consisting of DMEM supplemented with 10% fetal bovine serum, non-essential amino acids, L-glutamine, and penicillin/streptomycin.
- 53BP1-mCherry (N-Myc-53BP1 WT pLPC-Puro; Addgene plasmid 19836) and H2B-GFP (pCLNR-H2BG; Addgene plasmid 17735) fusion gene constructs also expressing puromycin or G418 resistance genes, respectively, were transduced (fibroblasts) or transfected (HEK293) using SuperFect (Qiagene) into cells and maintained under drug selection. Maintenance of fluorescence was periodically checked.
- 24-48 hr before image acquisition, cells were trypsinized into a single-cell suspension and seeded at a low density onto 3.5-cm FluroDish glass bottom plates.
B. Microscope Setup and Image Acquisition
This protocol was developed using the Zeiss Cell Observer SD spinning disk confocal microscope equipped with an AxioObserver Z1 stand, a dual-channel Yokagawa CSU-X1A 5000 spinning disk unit, 2 Photometrics QuantEM 512SC emCCD cameras, an HXP 120C fiber-based illuminator, 4 lasers (a Lasos 100mW multi-line Argon [458, 488, 514 nm], 50 mW 405 nm diode, 40 mW 561 nm diode, and 30 mW 635 nm diode), AOTF, a Pecon XL multiS1 stage incubation system, Zeiss incubation modules (O2 Module S, CO2 Module S, TempModule S, Heating Unit XL S) and a Prior motorized XY stage with a NanoScanZ piezo Z insert. To minimize spherical aberrations while imaging live cells supported in an aqueous medium, a C-Apochromat 63x/1.20 Water/Corr objective lens and Zeiss Immersol W immersion fluid (with a refractive index n = 1.334) were used. For 2 channel confocal imaging, a RQFT 405/488/568/647 dichroic mirror and BP525/50 (green) and BP629/62 (red) emission filters were used. The system software used was Zeiss Axiovision (ver. 4.8.2.0) with AV4 Multi-channel/Z/T, Fast Imaging, Physiology, MosaiX, Mark & Find, Dual Camera, Inside 4D, Autofocus and 3-D Deconvolution modules.
- Ensure that CO2 gas is running to the CO2 Module of the incubation system. If the microscope is supported by an anti-vibration air table, turn on the air supply (or N2 gas) for the air table.
- Turn on the power for the microscope stand, spinning disk unit, cameras, incubation modules, HXP illuminator, motorized stage, Argon laser, and computer.
- After 1 min of warm-up time, turn the ignition key for the Argon laser to "On".
- On the Zeiss laser control panel, turn on the switches for the laser lines to be used.
- Switch the toggle switch for the Lasos Argon laser controller from "standby" to "laser run" and adjust the light controller to the optimal level (i.e. just below the point where the green indicator turns red).
- Start up the AxioVision software. Note: the user interface for AxioVision may be customized with windows and pull-down menus that are specific for the particular microscope and components that it controls. As such, each system has a potentially unique interface. Therefore, generic instructions for software manipulation are provided in subsequent steps, rather than directions for specific software windows, tabs and/or pull-down menus.
- Approximately 1 hr prior to imaging, in the software, locate the controls for the incubator and turn on the heating for the upper chamber and the stage plate. Set the temperature to 37 °C. Turn on the CO2 control and set the level at 5%.
- Select the objective lens for imaging. In this study, a 63x/1.20 N.A. C-Apochromat Water/Corr objective lens was used. Note: For this lens, an immersion medium with a refractive index similar to water (Zeiss Immersol W immersion fluid) is needed.
- If multiple separate positions are to be sampled over a long period of time, be certain to apply a sufficient amount of immersion medium to ensure that it is carried from one position to another.
- Place the dish on the stage and bring the objective lens up into contact with the bottom.
- Using either the microscope controls or the software, direct the emitted light to the eyepieces and select the appropriate widefield filter set for the fluorescent signal of interest (for this study, "Red" filter set for 53BP1 and a "GFP" filter set for H2B).
- View through the ocular lenses, focus the image, and locate a suitable field of cells.
- Using either the software or the microscope controls, direct the emitted light away from the eyepieces to the port with the confocal spinning disc unit.
- In the software, turn on the appropriate laser (for this study, the 561 nm laser for 53BP1 and the 488 nm line of the Argon laser for H2B).
- For each channel, adjust the intensity of the laser by adjusting the acousto-optic tunable filter (AOTF) control to an appropriate level.
- Select the appropriate dichroic mirror (RQFT 405/488/568/647) and emission filters (BP 629/62 for 53BP1 and BP 525/50 for H2B). Open the shutter to the spinning disc unit.
- Select the "Live" window to display the current field of view.
- In the software, open the "Camera" control, select the camera to be used (if a dual camera system) and set exposure time to approximately 100 ms. Adjust the % and EM gain as necessary. Note: The 53BP1-mCherry construct appears somewhat dim. We found it helpful to increase the EM gain.
- In the software, open the control for the confocal spinning disc unit and adjust the spinning disc speed by entering the camera exposure time that was set to capture a suitable image (e.g. ~100 ms). Click "Set" to lock in the change.
- Open "multi-dimensional acquisition". Select the channel tab and load/select appropriate channels. For this study, we're using channels defined for dsRed (561 nm laser excitation and BP 629/62 filter for emission) and GFP (488 nm laser line for excitation and BP525/50 filter for emission).
- To ensure image registration, a common dichroic mirror (RQFT 405/488/568/647) was used for both channels. Set the software to "Autofocus". Note: Go to Tools→ Settings editor to adjust the MDA settings. Ensure that adequate laser power is set ("Adequate laser power" refers to a setting which enables you to excite the appropriate fluorophore while minimizing photo-bleaching).
- In the MDA window, select the z-stack tab. Select "Z-stack at current focus position". Set the range for ~10 μm z-stack and choose "optimal" for the number of steps to ensure Nyquist sampling through Z. Note: The exact size of the z-stack will depend on the height of your cells. Adjust accordingly.
- 23. Click "Start" and analyze the resulting z-stack image to ensure the settings are appropriate for what you are investigating (e.g. in this study, it was important to image the entire nucleus).
- Select the "T" (time) tab. For our experiment with camptothecin, we set the interval between imaging timepoints to 5 min and the overall duration of the session for 1 hr for a control (non-treated cells) video. Note: to minimize photo-bleaching of the cells, the experiment should be configured such that the AOTF blanks the laser between imaging time points.
- For multi-point imaging of several cells in the dish, in the MDA window, select the position tab. Ensure "Apply 'setting before/after time point' per position" is checked. Select "Mark_Find". Using the "Live" view, move the dish around and select appropriate fields of view.
- Click "Start" in the multi-dimensional-acquisition menu to begin the experiment and record a control video (of untreated cells).
- After the control video, add the appropriate treatment (in this case, 10 μM camptothecin). We recorded the experimental (drug-treated cells) video at 5-10 min intervals for 2-4 hr. Note: An important issue to consider is the speed with which a given effect occurs after treatment. As seen in the video, 53BP1 foci begin to from ~5 min after addition of CPT. Though a relatively easy process, adding drug to the dish manually and re-calibrating the microscope does take time. Consideration must be given to this when designing experiments.
- For monitoring mitosis, we set the interval to 7.5 min and recorded for 4-5 hr (the specific settings depend upon the cell line used and the length of its cell cycle). Adjustments may need to be made to prevent photo-bleaching during longer recordings.
C. Image Processing and Analysis
This protocol was developed using Volocity software (PerkinElmer). The software used to acquire this data (AxioVision) also has the ability to process and analyze images. Users are encouraged to utilize the software available to them, and consult appropriate literature regarding their application.
- Open Volocity software. Create and name a new library, and import your video files.
- For viewing the files, we usually find it most helpful to use the "Extended Focus" setting. This superimposes the z-stack slices and, for this protocol, allows us to visualize 53BP1 foci in different areas of the nucleus.
- Adjust videos as necessary. Volocity is equipped with a range of tools to improve the quality of acquired images. By ensuring the settings on the microscope were appropriate, much time can be saved in editing later on. Often, it is helpful to deconvolve your images, and adjust the brightness/contrast. Your specific editing needs will vary based on the experiment.
- Add a relative time stamp and scale bar.
- To view cells in 3-D, switch to the "3-D Opacity" setting. This permits rotation of the 3-D rendered cells in space, thus providing multiple perspectives of structures of interest within the cells. Note: It is helpful to visualize cells in different planes to determine where structures of interest travel. For example, in the "Extended Focus" setting it is difficult to discern that the 53BP1 does, in fact, dissociate from DNA during mitosis. However, this is readily apparent in 3-D.
- Movies and still images may be exported into a variety of file types based on user preference.
D. Representative Results
An example of 53BP1 foci formation in response to CPT is shown in Figure 1. Cells exposed to CPT form foci within 5-10 min, and maintain these foci throughout the duration of recording. As shown in Figure 2, 53BP1 dissociates from chromatin at the onset of mitosis, forming a thin haze around the condensing chromosomes. As telophase occurs and mitosis comes to an end, 53BP1 once again aggregates into distinct foci. While the HEK293 cells were not exposed to CPT, they nevertheless formed abundant, spontaneous 53BP1 repair foci generated by endogenous DNA damage. This observation allowed us to conclude that 53BP1 does not form foci during early mitosis, in line with a previous report showing a similar effect after the exposure of cells to ionizing radiation and radiomimetic drugs 5.

Figure 1. Camptothecin (10 μM) causes DSBs and 53BP1 foci formation in cycling fibroblasts within 30 min of adding the drug to the medium.
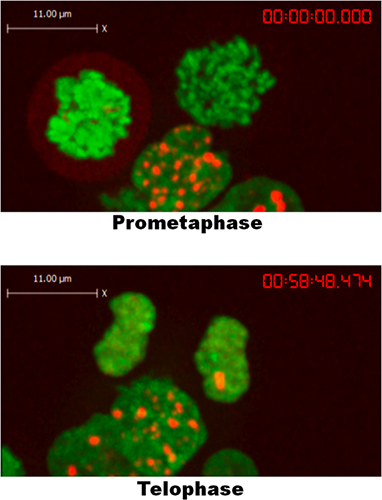
Figure 2. 53BP1 does not form foci during mitosis until telophase/G1 in HEK293 cells.
Discussion
Maintenance of genomic integrity is crucial for cell survival. Failure to preserve the genome results in premature aging, carcinogenesis, or death 8. There is intense interest in discerning how the DDR functions, stemming from its importance to both basic and clinical research. Many techniques have been developed over the years to aid in the study of how cells detect and repair DNA damage. Traditional methods such as immunocytochemistry and western blotting have been mainstays of the field, though recen...
Disclosures
No conflicts of interest declared.
Acknowledgements
Supported in part by R01NS064593 and R21ES016636 (K.V.). Microscopy was performed at the VCU - Department of Neurobiology & Anatomy Microscopy Facility, supported, in part, with funding from NIH-NINDS Center core grant 5P30NS047463. The spinning disc confocal microscope was purchased with an NIH-NCRR award (1S10RR027957).
Materials
| Name | Company | Catalog Number | Comments |
| CellStar culture dishes | Greiner Bio-one | ||
| FluroDish glass bottom dishes | World Precision Instruments, Inc. | ||
| MEM media | GIBCO | ||
| Non-essential amino acids | GIBCO | ||
| Amino acids | GIBCO | ||
| Vitamins | GIBCO | ||
| Sodium Pyruvate | Invitrogen | ||
| Penicillin/Streptomycin | HyClone | ||
| Fetal Bovine Serum | GIBCO | ||
| N-Myc-53BP1 WT pLPC-Puro; plasmid 19836 | Addgene | ||
| pCLNR-H2BG; plasmid 17735 | Addgene | ||
| SuperFect | Qiagen | ||
| Zeiss Cell Observer SD Imaging system | Zeiss | ||
| AxioVision (release 4.8.2) | Zeiss | ||
| Zeiss Immersol W Oil | Zeiss | ||
| Volocity software (version 6.0) | PerkinElmer |
References
- Botuyan, M. V., Lee, J., Ward, I. M., Kim, J. E., Thompson, J. R., Chen, J., Mer, G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 127, 1361-1373 (2006).
- Dimitrova, N., Chen, Y. C., Spector, D. L., de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 456, 524-528 (2008).
- Feuerhahn, S., Egly, J. M. Tools to study DNA repair: what's in the box. Trends Genet. 24, 467-474 (2008).
- Giunta, S., Belotserkovskaya, R., Jackson, S. P. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 190, 197-207 (2010).
- Giunta, S., Jackson, S. P. Give me a break, but not in mitosis: the mitotic DNA damage response marks DNA double-strand breaks with early signaling events. Cell Cycle. 10, 1215-1221 (2011).
- Golding, S. E., Morgan, R. N., Adams, B. R., Hawkins, A. J., Povirk, L. F., Valerie, K. Pro-survival AKT and ERK signaling from EGFR and mutant EGFRvIII enhances DNA double-strand break repair in human glioma cells. Cancer Biol. Ther. 8, 730-738 (2009).
- Huyen, Y., Zgheib, O., Ditullio, R. A., Gorgoulis, V. G., Zacharatos, P., Petty, T. J., Sheston, E. A., Mellert, H. S., Stavridi, E. S., Halazonetis, T. D. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 432, 406-411 (2004).
- Jackson, S. P., Bartek, J. The DNA-damage response in human biology and disease. Nature. 461, 1071-1078 (2009).
- Kanda, T., Sullivan, K. F., Wahl, G. M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377-385 (1998).
- Massignani, M., Canton, I., Sun, T., Hearnden, V., Macneil, S., Blanazs, A., Armes, S. P., Lewis, A., Battaglia, G. Enhanced fluorescence imaging of live cells by effective cytosolic delivery of probes. PLoS One. 5, e10459 (2010).
- Nakamura, K., Sakai, W., Kawamoto, T., Bree, R. T., Lowndes, N. F., Takeda, S., Taniguchi, Y. Genetic dissection of vertebrate 53BP1: a major role in non-homologous end joining of DNA double strand breaks. DNA Repair (Amst). 5, 741-749 (2006).
- Schultz, L. B., Chehab, N. H., Malikzay, A., Halazonetis, T. D. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell. Biol. 151, 1381-1390 (2000).
- Valerie, K., Povirk, L. F. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 22, 5792-5812 (2003).
- Wang, B., Matsuoka, S., Carpenter, P. B., Elledge, S. J. 53BP1, a mediator of the DNA damage checkpoint. Science. 298, 1435-1438 (2002).
- Ward, I. M., Minn, K., Jorda, K. G., Chen, J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278, 19579-19582 (2003).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved