A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Iterative Optimization of DNA Duplexes for Crystallization of SeqA-DNA Complexes
In This Article
Summary
Crystal structure of protein–DNA complexes can provide insight into protein function, mechanism, as well as, the nature of the specific interaction. Here, we report how to optimize the length, sequence and ends of duplex DNA for co-crystallization with Escherichia coli SeqA, a negative regulator of replication initiation.
Abstract
Escherichia coli SeqA is a negative regulator of DNA replication that prevents premature reinitiation events by sequestering hemimethylated GATC clusters within the origin of replication1. Beyond the origin, SeqA is found at the replication forks, where it organizes newly replicated DNA into higher ordered structures2. SeqA associates only weakly with single GATC sequences, but it forms high affinity complexes with DNA duplexes containing multiple GATC sites. The minimal functional and structural unit of SeqA is a dimer, thereby explaining the requirement of at least two GATC sequences to form a high-affinity complex with hemimethylated DNA3. Additionally, the SeqA architecture, with the oligomerization and DNA-binding domains separated by a flexible linker, allows binding to GATC repeats separated by up to three helical turns. Therefore, understanding the function of SeqA at a molecular level requires the structural analysis of SeqA bound to multiple GATC sequences. In protein-DNA crystallization, DNA can have none to an exceptional effect on the packing interactions depending on the relative sizes and architecture of the protein and the DNA. If the protein is larger than the DNA or footprints most of the DNA, the crystal packing is primarily mediated by protein-protein interactions. Conversely, when the protein is the same size or smaller than the DNA or it only covers a fraction of the DNA, DNA-DNA and DNA-protein interactions dominate crystal packing. Therefore, crystallization of protein-DNA complexes requires the systematic screening of DNA length4 and DNA ends (blunt or overhang)5-7. In this report, we describe how to design, optimize, purify and crystallize hemimethylated DNA duplexes containing tandem GATC repeats in complex with a dimeric variant of SeqA (SeqAΔ(41-59)-A25R) to obtain crystals suitable for structure determination.
Protocol
1. Protein Purification
The flexible linker connecting the N- (oligomerization) and C-terminal (DNA binding) domains of SeqA aids the recognition of hemimethylated GATC repeats separated by one to three turns on the DNA. For this study, we used a dimeric variant of SeqA (SeqAΔ(41-59)-A25R) with a point mutation in the N-terminal domain that prevents further oligomerization and a shortened linker that restricts DNA binding to tandem GATC repeats separated exclusively by one turn on the DNA (Figure 1)2,8.
- Transform BL21 (DE3) cells with the plasmid encoding SeqA under the control of T7 promoter,
- Plate the transformation reaction in LB-agar plates including 100 μg/ml ampicillin,
- Pick mixed colonies to inoculate a small-scale overnight culture (LB media with 100 μg/ml ampicillin),
- The next morning inoculate a 1 L of media using a 1:100 dilution of the overnight culture,
- Grow the cells to an OD600 of ~0.7 and induce protein production by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM,
- Continue the incubation for 3 hr at 37 °C with orbital shaking and then harvest the cells by centrifugation (10 min at 3,300 g),
- Resuspend the cell pellet in purification buffer A and lyse by sonication,
- Clear the lysate by centrifugation (40 min at 39,000 g) and load the supernatant onto a heparin column equilibrated with purification buffer,
- Elute SeqA using a linear gradient to 1 M NaCl (SeqA elutes at ~0.7 M NaCl),
- Pool the SeqA-containing fractions together, dilute to lower the ionic strength of the sample and load into a cation-exchange chromatography column equilibrated with purification buffer,
- Using a linear salt gradient, pure SeqA elutes at ~0.4 M NaCl,
- Pool the SeqA-containing fractions together, concentrate (3 mg/ml) and store in storage buffer.
2. DNA Purification
- Order complementary unmethylated and methylated oligonucleotides from your favorite company,
- Dissolve 1 μmol of each lyophilized single strand DNA in 800 μl of autoclaved ddH2O, vortex and let it sit for 10-20 min,
- Add 800 μl of preheated 2X loading buffer to each oligonucleotide,
- For 20-30 nucleotides long oligonucleotides, prepare a large 10% denaturing gel (160 x 250 x 1 mm):
* Mix well 80 ml of 10% PAGE mix, 80 μl TEMED and 800 μl ammonium persulfate per gel and pour,
* Once polymerized, remove the comb and rinse the wells with ddH2O thoroughly,
* Assemble the gels on gel cast including cooling plate and then fill the top and bottom reservoirs with running buffer (1X TBE),
* Pre-run the gel at 700-750 V to warm the gel up to 55 °C,
* Stop the run and rinse the wells thoroughly with running buffer. - Heat the oligonucleotides to 90 °C for 2 min,
- Vortex and spin the samples and immediately before loading the gel,
- Run the gel at ~700 V and stop it once your oligonucleotide has migrated halfway. (Note that on a 10% polyacrylamide gel bromophenol blue co-migrates with oligonucleotides ~20 bases long and xylene cyanol FF with ~60 bases long),
- Stop the gel, disassemble it from the gel box and remove the spacers,
- On a flat surface, remove one glass plate and cover the gel with plastic wrap,
- Turn the gel around, remove the other glass plate and cover it with plastic wrap,
- Mark the bands using UV light and a fluorescent plate behind the gel to see the DNA shadow,
- Cut the band with a razor blade into small pieces and transfer them into a sterile 15 ml tube,
- Add 9 ml of elution buffer and elute overnight at 37 °C with agitation,
- Carefully transfer the solution to an autoclaved centrifuge tube using a gel-loading pipette tip to avoid transferring acrylamide pieces and add 1 ml of 3 M sodium acetate at pH 7 (1:10 dilution) plus 25 ml of chilled 100% ethanol (2.5 volumes),
- Incubate at -20 °C for at least 3 hr,
- Spin down and transfer the supernatant to a separate tube,
- Dry the pellet on the speed-vac at medium heat,
- Resuspend the pellet in 400 μl of autoclaved ddH2O and transfer to a fresh tube,
- Add 40 μl of 3 M sodium acetate at pH 7 and 1 ml of 100% ethanol; mix well (vortex) and incubate 30 min at room temperature followed by 30 min at -20 °C,
- Spin for 15 min at 18,000 g and discard the supernatant,
- Rinse the pellet with 100 μl of 70% cold ethanol to remove residual salt from the pellet and spin for 6 min at 18,000 g. Discard the ethanol and dry the pellet on speed-vac,
- Resuspend the pellet in a total of 100 μl of autoclaved ddH2O. Measure the concentration of the oligonucleotide,
- To anneal the hemimethylated DNA duplexes, mix equimolar concentrations of the complementary single strands and heat the mixtures to 95 °C in a water bath for 5 min and then let them cool down slowly to room temperature inside the water bath.
3. Protein-DNA Complex Formation and Analysis
- Mix equal volumes of purified SeqAΔ(41-59)-A25 (81 μM) and hemimethylated DNA (81 μM),
- Incubate at room temperature for 15 min and store at 4 °C until you are ready to use it,
- Screen for crystallization conditions using commercial sparse-matrix screens,
- Once initial crystallization leads have been identified, optimize the conditions to grow diffraction quality crystals,
- Cryoprotect the resulting SeqA-DNA crystals by either increasing the amount of PEG 400 present in the crystallization solution to a final concentration of 25% (v/v) or adding 20% glycerol (v/v) to the crystallization solution,
- Scoop individual crystals with a nylon loop, and flash-freeze them in liquid nitrogen,
- Test the diffraction limit of each crystal at 100 K.
4. Representative Results
To obtain the crystal structure of SeqAΔ(41-59)-A25R bound to hemimethylated DNA, we consecutively optimized three parameters on the DNA: (i) separation between hemimethylated GATC sequences; (ii) length of the duplex; and (iii) the absence/presence of 5' overhangs.
Electro-mobility shift assays indicate that SeqAΔ(41-59)-A25R preferentially binds GATC repeats separated by 9-10 base pairs (Figure 1). Therefore, we initially screened duplexes 23-24 base pairs (bps) long containing two hemimethylated GATC sequences separated by either 9 or 10 bps. Three duplexes yielded nicely shaped crystals (Figure 2). Although none of the crystals diffracted to high resolution, the 23 bps long duplex with the two GATC sites separated by 9 bps diffracted X-rays better than the rest, indicating that a GATC separation of 9 bps was preferred for crystallization. Therefore, we fixed the inter-GATC spacing to 9 bps for all subsequent screens.
Generally, DNA crystallization is favored for duplex lengths corresponding to exact helical turns because multiple DNA molecules can stack head-to-tail to form a continuous B-DNA within the crystal9. Therefore, we shortened the overall length of the duplexes to 21 bps (i.e. two helical turns). While a 21 bps duplex with blunt ends did not yield diffraction quality crystals (data not shown), a 21 bps duplex with a single 5' overhang nucleotide on each end did yield crystals that diffracted to 5 Å in our home source. The improvement on the diffraction limit suggested that end-to-end duplex association was indeed favoring crystal packing.
Since SeqA interacts with GATC sequences that are on the same face of the DNA, the opposite face of the DNA duplex should be exposed to the solvent. To further improve the crystals of the complex, we then modified the sequence of the duplex to include a CG dinucleotide between the two GATC sites to promote groove-backbone interactions with adjacent DNA molecules in the crystal through the opposite face of the duplex, a method that has been used to enhance DNA crystallization in the past10. However, the diffraction limit of the crystals grown with the CG-containing duplex was identical to those grown with a similar DNA duplex that did not contain the CG dinucleotide (Figure 3). This result indicated that groove-backbone interactions were not important in this case. We subsequently optimized the length of the overhangs by comparing the crystals grown with a DNA duplex that had two additional nucleotides at each 5' end. This change had a drastic effect on crystal morphology, as well as, diffraction limit indicating that the additional nucleotide dramatically changed the molecular contacts and enhanced crystal organization (Figure 3).
The crystal structure of SeqAΔ(41-59)-A25R bound to this last DNA duplex confirmed that the free face of the DNA duplex does not engage in crystal contacts, as expected from the limited effect of introducing a CG-dinucleotide. Despite the relative size of protein and DNA, most interactions between symmetry mates are mediated by protein-protein and protein-DNA interactions (Figure 4). Interestingly, in this particular case, the beneficial effect of a 5' dinucleotide overhang is not due to the formation of a pseudo-continuous DNA. Instead, the 5' end of the methylated strand projects away from the DNA axes and interacts with the proximal SeqA molecule of the complex, explaining why two nucleotides were strictly required to change the crystal packing8,11.
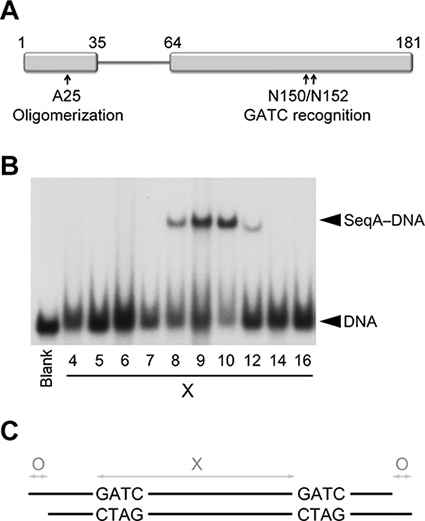
Figure 1. Binding of SeqAΔ(41-59)-A25R to hemimethylated DNA. (A) Schematic diagram of the domains of SeqA that mediate protein oligomerization and DNA binding. (B) Electrophoretic mobility shift assay of SeqAΔ(41-59)-A25R with DNAs containing two hemimethylated GATC sequences separated by an increasing number of base pairs (X). The left-most lane (labeled Blank) contains an equimolar mixture of DNAs with 5, 7, 12, 21, 25 and 34 base pairs between the two GATC sequences in the absence of SeqAΔ(41-59)-A25R. (C) Diagram depicting the three variables optimized on the DNA duplexes to obtain diffraction quality crystals.

Figure 2. Effect of varying the inter-GATC distance. Summary of the oligonucleotides used and crystals obtained with 23-24 bp long duplexes containing two hemimethylated GATC sites separated by 9 or 10 base pairs. All pictures of crystals were taken at the same magnification and the scale bar indicates 100 μm. The resolution limit of each SeqAΔ(41-59)-A25R-DNA crystal is based on diffraction images collected on a Rigaku RU-300 X-ray generator system. Click here to view larger figure.

Figure 3. Effect of varying the DNA ends. Summary of the oligonucleotides used and crystals obtained with 21 bps long duplexes containing two hemimethylated GATC sites separated by 9 base pairs and including 0, 1 or 2 nucleotide 5' end overhangs. All pictures of crystals were taken at the same magnification and the scale bar indicates 100 μm. The resolution limit of each SeqAΔ(41-59)-A25R-DNA crystal is based on diffraction images collected at beamlines X12C and X29 (NSLS, BNL). Click here to view larger figure.
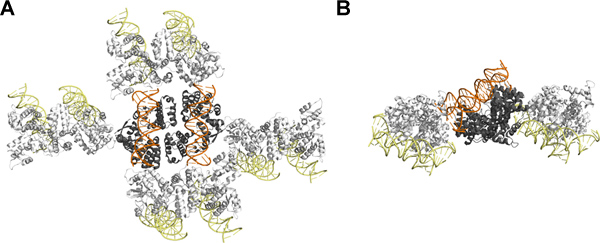
Figure 4. Crystal packing of SeqAΔ(41-59)-A25R bound to DNA with dinucleotide overhang: Viewed from the (A) top and the side (B). The asymmetric unit contains two SeqAΔ(41-59)-A25R-DNA complexes where the protein is shown in grey while the DNA is shown in orange. Symmetry related SeqAΔ(41-59)-A25R-DNA molecules are shown in white for the protein and yellow for the DNA. This figure was prepared using PyMOL12. This figure is related to Movie 1. Click here to view larger figure.
Movie 1. Click here to view movie.
Discussion
One of the biggest challenges in macromolecular X-ray crystallography is obtaining diffraction quality crystals. In the case of protein or protein-DNA complexes, this challenge is exacerbated due to the additional variables that must be optimized. It is widely believed that the length of the DNA and the presence of sticky overhangs to enhance association of neighboring DNA molecules in a longer pseudo-duplex are the main parameters to optimize. However, we have shown that the nature and length of these overhangs can have...
Disclosures
No conflicts of interest declared.
Acknowledgements
The authors wish to thank the PXRR staff at the NSLS (Brookhaven National Laboratory) for assistance during data collection and Monica Pillon for help with DNA purification. This work was supported by the Canadian Institutes of Health Research (MOP 67189).
Materials
| Name | Company | Catalog Number | Comments |
| TRIS | Bioshop | TRS003.5 | |
| Ethylenediaminetetraacetic acid (EDTA) | Fisher Scientific | E478-500 | |
| Dithiotheitrol (DTT) | Bio Basic Inc. | DB0058 | |
| NaCl | Bioshop | SOD002.10 | |
| Glycerol | Caledon | 5350-1 | |
| Sucrose | Sigma-Aldrich | S5016-500G | |
| Sodium dodecyl sulfate (SDS) | Bioshop | SDS001.500 | |
| Urea | Bioshop | URE001.5 | |
| 40% 29:1 Bis/acrylamide | Bio Basic Inc. | A0007-500ml | Store at 4 °C |
| Boric acid | EMD | BX0865-1 | |
| Xylene cyanol FF | Bio-Rad | 161-0423 | |
| Bromophenol Blue | Bioshop | BR0222 | |
| Dual Adjustable Vertical Gel System | C.B.C. Scientific Company Inc. | DASG-250 | |
| Index crystallization screen | Hampton Research | HR2-144 | Store at 4 °C |
| Wizard I crystallization screen | Emerald BioSystems | EBS-WIZ-1 | Store at 4 °C |
| Wizard II crystallization screen | Emerald BioSystems | EBS-WIZ-2 | Store at 4 °C |
| Classics crystallization screen | Qiagen | 130701 | Store at 4 °C |
| Intelliplate trays | Art Robbins Instruments | 102-0001-00 | |
Solutions Protein purification buffer: 100 mM TRIS pH 8, 2 mM EDTA, 2 mM DTT and 5% glycerol. Protein storage buffer: 20 mM TRIS pH 8, 150 mM NaCl, 5 mM DTT, 0.5 mM EDTA and 5% glycerol. Gel loading mix: Add 20 g of sucrose, 25 mg of bromophenol blue, 25 mg of xylene cyanol FF, 1 ml of 10% w/v SDS and 10 ml of 10X TBE to 70 ml of autoclaved ddH2O. Stir with mild heating until sucrose is dissolved and adjust the final volume to 100 ml with autoclaved ddH2O. Store at 4 °C. 2X loading buffer: Add 11 g of urea to 10 ml of gel loading mix. Stir on a hot plate until urea dissolves. Aliquot in 2 ml tubes and store at 4 °C. 10X PAGE mix: Mix 420.4 g of urea, 100 ml of 10X TBE (autoclaved), 250 ml of 40% 29:1 Bis/Acrylamide in ddH2O. Stir until totally dissolved and adjust volume to 1 liter. Store in dark bottles at 4 °C. 10X TBE: Dissolve 108 g of TRIS, 55 g of boric acid and 9.3 g of EDTA in 1 liter of ddH2O. Autoclave and store at room temperature. Elution buffer: Dilute 8 ml of 5 M NaCl, 2 ml of 1 M TRIS pH 7.5, 0.4 ml of 0.5 M EDTA pH 8 on 200 ml of ddH2O. Autoclave and store at room temperature. | |||
References
- Campbell, J. L., Kleckner, N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 62, 967-979 (1990).
- Guarné, A. Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization. Embo. J. 24, 1502-1511 (2005).
- Brendler, T., Austin, S. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. Embo. J. 18, 2304-2310 (1999).
- Jordan, S. R., Whitcombe, T. V., Berg, J. M., Pabo, C. O. Systematic variation in DNA length yields highly ordered repressor-operator cocrystals. Science. 230, 1383-1385 (1985).
- Tan, S., Hunziker, Y., Pellegrini, L., Richmond, T. J. Crystallization of the yeast MATalpha2/MCM1/DNA ternary complex: general methods and principles for protein/DNA cocrystallization. J. Mol. Biol. 297, 947-959 (2000).
- Rice, P. A., Yang, S., Mizuuchi, K., Nash, H. A. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 87, 1295-1306 (1996).
- Yang, W., Steitz, T. A. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 82, 193-207 (1995).
- Chung, Y. S., Brendler, T., Austin, S., Guarne, A. Structural insights into the cooperative binding of SeqA to a tandem GATC repeat. Nucleic Acids Res. , (2009).
- Anderson, J., Ptashne, M., Harrison, S. C. Cocrystals of the DNA-binding domain of phage 434 repressor and a synthetic phage 434 operator. Proc. Natl. Acad. Sci. U.S.A. 81, 1307-1311 (1984).
- Timsit, Y., Moras, D. DNA self-fitting: the double helix directs the geometry of its supramolecular assembly. Embo J. 13, 2737-2746 (1994).
- Chung, Y. S., Guarne, A. Crystallization and preliminary X-ray diffraction analysis of SeqA bound to a pair of hemimethylated GATC sites. Acta Crystallogr Sect F Struct Biol Cryst Commun. 64, 567-571 (2008).
- DeLano, W. L. . The PyMOL Molecular Graphic Systems. , (2002).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved