A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Preparation of Cell-lines for Conditional Knockdown of Gene Expression and Measurement of the Knockdown Effects on E4orf4-Induced Cell Death
In This Article
Summary
Contribution of the ACF chromatin remodeling factor to E4orf4-induced cell death was measured. The protocol includes selection of cell clones in which doxycycline treatment induces conditional knockdown of the ACF subunits Acf1 and SNF2h, and use of the DAPI assay to measure E4orf4-induced cell death in the inducible cell lines.
Abstract
Functional inactivation of gene expression in mammalian cells is crucial for the study of the contribution of a protein of interest to various pathways1,2. However, conditional knockdown of gene expression is required in cases when constitutive knockdown is not tolerated by cells for a long period of time3-5. Here we describe a protocol for preparation of cell lines allowing conditional knockdown of subunits of the ACF chromatin remodeling factor. These cell lines facilitate the determination of the contribution of ACF to induction of cell death by the adenovirus E4orf4 protein6. Sequences encoding short hairpin RNAs for the Acf1 and SNF2h subunits of the ACF chromatin remodeling factor were cloned next to a doxycycline-inducible promoter in a plasmid also containing a gene for the neomycin resistance gene. Neomycin-resistant cell clones were selected in the presence of G418 and isolated. The resulting cell lines were induced by doxycycline treatment, and once Acf1 or SNF2h expression levels were reduced, the cells were transfected with a plasmid encoding E4orf4 or an empty vector. To confirm the specific effect of the shRNA constructs, Acf1 or SNF2h protein levels were restored to WT levels by cotransfection with a plasmid expressing Acf1 or SNF2h which were rendered resistant to the shRNA by introduction of silent mutations. The ability of E4orf4 to induce cell death in the various samples was determined by a DAPI assay, in which the frequency of appearance of nuclei with apoptotic morphologies in the transfected cell population was measured7-9.
The protocol described here can be utilized for determination of the functional contribution of various proteins to induction of cell death by their protein partners in cases when constitutive knockdown may be cell lethal.
Protocol
1. Generation of Inducible Cell Lines
- Prior to the experiment one needs to find the minimal concentration of the drug G418 which kills all the cells used for preparation of the desired cell lines. For this purpose, plate the cells of choice in several duplicate plates at 70% confluency in the medium that will be used during the experiment and add increasing concentrations of G418 (0-1,000 μg per ml). Monitor the cells daily to determine the minimal G418 concentration that kills the cells efficiently. Cell death is observed within 3-7 days.
- Prior to generation of cell lines it is recommended to verify by transient transfection assays that the plasmid expressing the shRNA can reduce to some extent expression of the gene under study. This may be done in any cell line that can be efficiently transfected.
- Plate T-REx-293 cells at a density of around 5x106 cells per 10 cm plate in 8 ml of DMEM medium (containing 10% Tet system approved FBS, 2mM L-glutamine, 100 units per ml penicillin and 0.1 mg per ml streptomycin, 5 μg per ml blasticidin). Incubate the plates overnight at 37 °C, 5% CO2.
- On the following day, change the medium to 8 ml fresh medium prior to transfection.
- Transfect the cells using 10 μg of pSuperior.neo+GFP plasmid (Figure 1) encoding Acf1 or SNF2h shRNA driven by a tetracycline-inducible H1 promoter, as well as the neomycin-resistance gene fused to GFP and driven by a constitutive PGK promoter. Add the DNA to 500 μl of 150 mM NaCl. Add the jetPIE reagent to another aliquot of 500 μl of 150 mM NaCl at 2 μl per μg DNA. Add the jetPIE solution to the DNA and mix well by vortexing. Incubate for 15 min at room temperature. Gently pipette DNA complexes onto the 10 cm plates containing 70-80% confluent cells. Return the plates to the incubator.
- The next day replace the medium with a selective medium (DMEM medium as described above containing an additional 500 μg per ml G418).
- For the next two weeks monitor the cells and replace the selective medium with a similar fresh medium every 3-4 days until colonies appear and can be visualized without a microscope.
- Prior to isolation of colonies, prepare 24-well plates with selective medium.
- Identify colonies by eye, and mark their location at the bottom of the plate using a colored marker. Verify under the microscope that the colonies are well separated.
- In the sterile hood, aspirate the medium from the plate, gently rinse with warm PBS and aspirate well all remaining liquid. Add 3 μl of 0.25 % trypsin/EDTA to one colony on the plate. Pipette the trypsin repeatedly until the cells detach and add them to one of the wells in the 24-well plate. Repeat this for several other colonies on the plate.
- Grow the cells at 37 °C, 5% CO2 until they fill the well. Then split the cells derived from each of the original colonies into three wells, each in a separate 12-well plate, and incubate overnight.
- On the following day, replace the medium in one plate with medium containing 1 μg per ml doxycycline from a stock solution prepared in double-distilled water (1-5 mg per ml). Replace the medium in a control plate with medium without doxycycline. Incubate the cells for 72 hr at 37 °C, 5% CO2.
- Harvest cells from one treated and one untreated well of each cell clone for preparation of protein extracts and Western blot analysis to determine the efficiency of Acf1 or SNF2h knockdown. Use antibodies to Acf1 or SNF2h as well as antibodies to alpha-Tubulin, serving as a loading control. Use the cells in the third well, left untreated, for expansion of selected cell clones in which doxycycline addition led to at least 50% reduction in the level of the protein under study. Freeze aliquots of these cells in 90% FBS, 10% DMSO for further use.
2. Induction of Knockdown and Transfection
- Plate cells in the selective medium in 10 cm plates. Add doxycycline at 1 μg per ml to half the plates and incubate at 37°C, 5% CO2 for 48-72 hr (see Figure 4).
- After 48-72 hr, trypsinize the cells from each group of cells, count them, and plate 1.5x106 cells per 6 cm plate in the same medium, with or without doxycycline. Incubate the cells at 37°C, 5% CO2 overnight. Plan so that each group of plates from section 2.1 will provide the cells for twelve 6 cm plates, including two duplicates for the DAPI assay for each point as well as one plate for Western blot analysis of each sample (see Figure 4).
- On the following day transfect the cells in 6 cm plates with the following combinations of plasmids: 1 μg of an empty plasmid or an identical quantity of the plasmid encoding E4orf4, together with 4 μg of a vector expressing Acf1-GFP or SNF2h-GFP rendered resistant to the shRNA in the cell clones by introduction of silent mutations, or 3 μg of the corresponding empty vector and 1 μg plasmid expressing GFP (Figure 4). Use the jetPIE reagent as described above (10 μl per 5 μg DNA) to prepare the transfection mix. For each sample, prepare a transfection mix for three plates.
- Following a 15 min incubation of DNA with the jetPIE reagent at room temperature, gently pipette equal amounts of DNA complexes from each transfection mix onto three 6 cm plates and incubate at 37°C, 5% CO2 overnight.
3. DAPI Assay in Transfected Cells
- The next day, extract proteins from one plate per sample for Western blot analysis to determine that Acf1 or SNF2h have been efficiently knocked down and that E4orf4 was expressed equally in the different samples.
- Aspirate the medium from the plates intended for the DAPI assay, wash the plates gently with PBS and add 1 ml of 4% paraformaldehyde prepared in PBS to cover the cells. Preparation of the paraformaldehyde solution and treatment of the cells with this chemical should be carried out in a chemical hood. Incubate for 15 min at room temperature without shaking.
- Aspirate the paraformaldehyde and wash three times with PBS, rocking at room temperature for 5 min each time. Add 80% ethanol kept at -20 °C and incubate at -20 °C for at least 1 hr. The plates can be kept under ethanol at -20 °C for several days as long as they are sealed well to prevent ethanol evaporation and drying.
- Aspirate the ethanol and wash the cells twice with PBS and once with PBS-BT (PBS containing 0.5% BSA and 0.05% tween-20), rocking at room temperature for 5 min each time.
- To prevent non-specific antibody binding, block the cells in 1 ml PBS-BT buffer containing 10% goat serum for 20 min while rocking at room temperature. Then wash the cells twice in PBS-BT, 5 min each wash.
- Incubate the cells with the primary antibody (an E4orf4-specific antibody in our case) in 1 ml PBS-BT for 1 hr while rocking at room temperature. Then wash twice in PBS-BT and once in PBS containing 0.1% BSA, 5 min each wash.
- Incubate the cells in 1 ml of PBS-0.1% BSA containing the appropriate secondary fluorescently-labeled antibody and 0.5 μg per ml final concentration of DAPI for 40 min while rocking at room temperature in the dark. Then wash with PBS for 5 min, dry well by aspiration and keep the plates upside down for an additional hour to overnight to achieve complete drying. Then mount cover slides on the cells, using Fluoromount-G solution. Keep plates at 4 °C in the dark until ready to count the apoptotic nuclei.
- Ask a colleague to identify the various plates by new numbers, so counting apoptotic nuclei in the various samples can be done in an unbiased way.
- Visualize the cells under an upright fluorescent microscope at a magnification of 400X (in air) or 630X (in immersion oil). Identify transfected cells which are stained for the various proteins expressed in each sample, and count the number of condensed or fragmented nuclei visualized by DAPI staining within the transfected cell population. Monitor several fields and count a total of at least 200 transfected cells.
- Calculate the percentage of nuclei with apoptotic morphology within the transfected cell population. Repeat the experiment, with duplicates, 2-3 times to calculate the statistical significance of changes between the various samples.
4. Representative Results
The efficiency of obtaining colonies in which conditional knockdown of gene expression has been successful is variable, depending on the gene involved. In our hands, we obtained 7 successful colonies out of 12 tested for Acf1 and 6 out of 18 for SNF2h. Figure 2 shows a representative plate of selected cell clones (Figure 2A) as well as a single colony (Figure 2B). Figure 3 shows a representative blot prepared with proteins extracted from cells of various clones grown in the presence or absence of doxycycline for 72 hr. The blot was stained with antibodies to SNF2h and to alpha-Tubulin, serving as a loading control. Two of the clones (number 4 and 5) showed a strong reduction in SNF2h levels upon doxycycline induction. It should be noted that although the pSuperior.neo+GFP plasmid (Figure 1) used for generation of the cell lines encodes a neo-GFP fusion protein, the green fluorescence in the stable cell lines was very low and expression of other GFP fusion proteins introduced transiently into these cells could be easily detected above the background green fluorescence.
A schematic representation of the experiments carried out in the cell clones to measure the effect of Acf1 or SNF2h knockdown on E4orf4-induced cell death is shown in Figure 4. The time required for knockdown of gene expression by doxycycline treatment can vary depending on the stability of the protein whose levels should be reduced. For Acf1 and SNF2h, a 72 hr treatment was required for efficient knockdown at the protein level.
Figure 5 shows an example of cells expressing E4orf4 and GFP and undergoing cell death manifested by the appearance of DAPI-stained nuclei with a condensed or fragmented morphology. Figure 6 shows the results of a representative experiment demonstrating that doxycycline-induced Acf1 knockdown led to an increase in E4orf4-stimulated cell death (Figure 6A). This increase did not result from an increase in E4orf4 levels (Figure 6B). Furthermore, restoration of Acf1 to the doxycycline-induced cells diminished the increase in E4orf4 toxicity to the levels observed in uninduced cells (Figure 6).
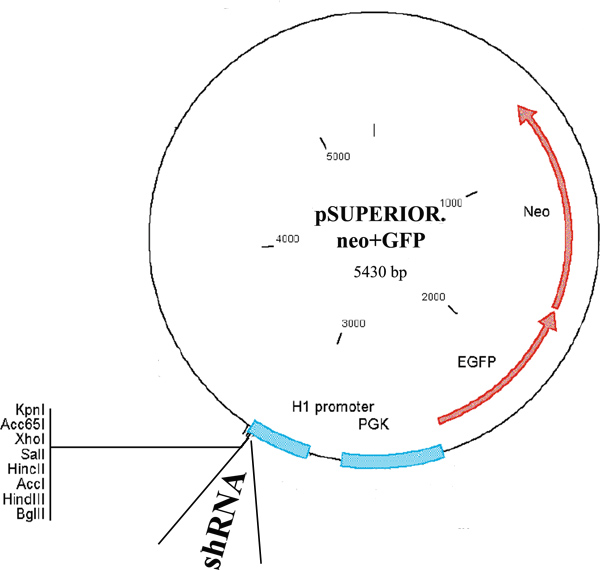
Figure 1. Map of the pSuperior.neo+GFP plasmid. The map shows the tetracycline-inducible H1 promoter driving shRNA expression and the PGK promoter driving expression of a neo-GFP fusion protein. The map was adapted from the OligoEngine pSuperior manual.
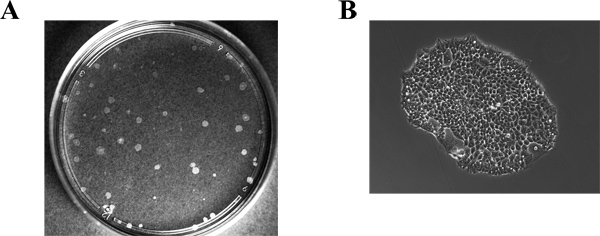
Figure 2. Identification of neomycin-resistant colonies. Images of a plate containing colonies (A) and of a single colony (B) are shown. The colonies were obtained by culturing the cells for 14 days in a selective medium containing neomycin.
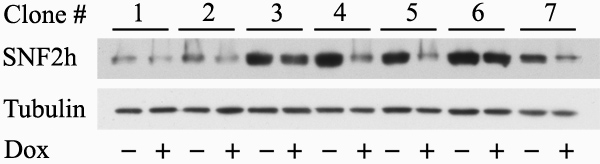
Figure 3. Examination of knockdown efficiency in selected colonies. Cells of several colonies selected in the presence of neomycin were plated in duplicate wells. One well of each sample was induced by doxycycline (+) and one well was left untreated (-). Proteins were extracted 72 hr post-induction and chromatographed on SDS-PAGE. The Western blot was stained successively with antibodies to SNF2h and to alpha-Tubulin (serving as a loading control) and shows SNF2h levels in induced and control cells.
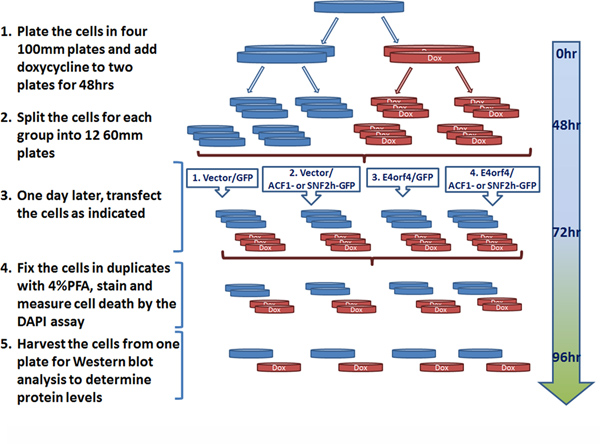
Figure 4. Planning a typical knockdown-DAPI assay experiment. Successive steps of the experiment are shown, including doxycycline treatment to achieve Acf1 or SNF2h knockdown, a transfection to restore Acf1 or SNF2h expression or to introduce an empty vector and to introduce E4orf4 or its corresponding empty vector into the cells, and analysis of the results. Doxycycline-treated plates are shown in red (Dox) and control plates are marked in blue. Click here to view larger figure.

Figure 5. Detection of E4orf4-induced cell death by the DAPI assay. Cells which underwent the various treatments described in Figure 4 were fixed and stained with E4orf4-specific antibodies and DAPI to visualize the nuclei of transfected cells. This specific picture was taken from a sample containing E4orf4 and the control GFP protein. (A) GFP. (B) E4orf4. (C) DAPI. (D) Merged images. The white arrows mark GFP- and E4orf4-transfected cells containing nuclei with apoptotic morphologies. Red arrows mark nuclei with irregular shapes which are not counted as apoptotic nuclei. Asterisks mark mitotic nuclei or nuclei that have just divided.
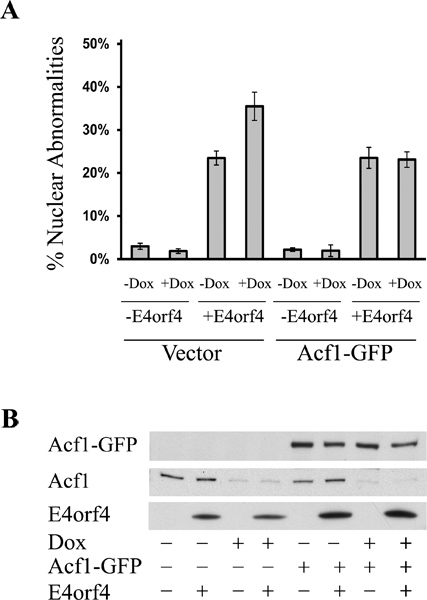
Figure 6. Acf1 knockdown enhances E4orf4-induced cell death. (A) Cells from the T-REx-293- derived cell line expressing Acf1-shRNA from a tetracycline-inducible promoter were induced with doxycycline (+Dox) or left untreated (-Dox). Three days later, the cells were transfected with plasmids expressing E4orf4 (+E4orf4) or an empty vector (-E4orf4) together with an empty vector or a plasmid expressing GFP-tagged Acf1, which was rendered resistant to the shRNA by the introduction of silent mutations. The cells were fixed twenty-four hours after transfection and stained with antibodies to E4orf4 and with DAPI. Induction of cell death was measured by the DAPI assay described above and the percentage of transfected cells with condensed or fragmented nuclei was determined. A representative experiment with three replicates is shown, and error bars represent the standard deviation. (B) Cells from parallel plates were harvested for Western blot analysis. The blot was stained with antibodies to E4orf4, GFP, and Acf1. The endogenous Acf1 and Acf1-GFP are shown in two separate panels.
Discussion
Knockdown of specific gene expression is an important approach to the investigation of the contribution of a protein to regulatory pathways. Since constitutive knockdown of expression of essential genes negatively affects cell proliferation, their study may require either transient introduction of siRNAs or application of stable conditional knockdown systems. Generation of stable cell lines with the capacity for drug-induced expression of shRNAs described here, provides an advantage over transient transfections wh...
Disclosures
No conflicts of interest declared.
Acknowledgements
This work was supported (in part) by THE ISRAEL SCIENCE FOUNDATION (Grant 399/11), by the Deutsche Forschungsgemeinschaft (DFG) within the framework of The German-Israeli Project Cooperation (DIP), and by the Rappaport Family Institute for Research in the Medical Sciences.
Materials
| Name | Company | Catalog Number | Comments |
| High glucose DMEM | Gibco | 41965 | |
| Tet system approved FBS | Clontech | 631106 | |
| L-glutamine | Gibco | 25030 | |
| Pen Strep | Gibco | 15140 | |
| 0.25% Trypsin-EDTA | Gibco | 25200 | |
| T-REx-293 cells | Invitrogen | R710-07 | |
| G418 | Sigma | A1720 | |
| blasticidin | Invitrogen | R210-01 | |
| pSuperior.neo+GFP plasmid | OligoEngine | VEC-PBS-0007/0008 | |
| jetPIE | Polyplus transfection | 101-40 | |
| doxycycline | Sigma | D9891 | |
| paraformaldehyde | Electron Microscopy Sciences | 15710 | |
| 4',6-diamidino-2-phenylindole (DAPI) | Sigma | D9542 | |
| Fluoromount-G | SouthernBiotech | 0100-01 |
References
- Brummelkamp, T. R., Bernards, R., Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science (New York, N.Y). 296, 550-553 (2002).
- Brummelkamp, T. R., Bernards, R., Agami, R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer. 2, 243-247 (2002).
- Czauderna, F. Inducible shRNA expression for application in a prostate cancer mouse model. Nucleic acids research. 31, e127 (2003).
- Matsukura, S., Jones, P. A., Takai, D. Establishment of conditional vectors for hairpin siRNA knockdowns. Nucleic acids research. 31, e77 (2003).
- Wiznerowicz, M., Trono, D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. Journal of. 77, 8957-8961 (2003).
- Brestovitsky, A., Sharf, R., Mittelman, K., Kleinberger, T. The adenovirus E4orf4 protein targets PP2A to the ACF chromatin-remodeling factor and induces cell death through regulation of SNF2h-containing complexes. Nucleic acids research. 39, 6414-6427 (2011).
- Livne, A., Shtrichman, R., Kleinberger, T. Caspase activation by adenovirus E4orf4 protein is cell line-specific and is mediated by the death receptor pathway. J. Virol. 75, 789-798 (2001).
- Shtrichman, R., Kleinberger, T. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J. Virol. 72, 2975-2982 (1998).
- Shtrichman, R., Sharf, R., Kleinberger, T. Adenovirus E4orf4 protein interacts with both Ba and B' subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Ba. Oncogene. 19, 3757-3765 (2000).
- Galluzzi, L. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell death and differentiation. 16, 1093-1107 (2009).
- Lavoie, J. N., Nguyen, M., Marcellus, R. C., Branton, P. E., Shore, G. C. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J. Cell Biol. 140, 637-645 (1998).
- Li, S. The adenovirus E4orf4 protein induces growth arrest and mitotic catastrophe in H1299 human lung carcinoma cells. Oncogene. 28, 390-400 (2009).
- Lavoie, J. N., Champagne, C., Gingras, M. -. C., Robert, A. Adenovirus E4 open reading frame 4-induced apoptosis involves dysregulation of Src family kinases. J. Cell Biol. 150, 1037-1055 (2000).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved