A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Transport Properties of Ibuprofen Encapsulated in Cyclodextrin Nanosponge Hydrogels: A Proton HR-MAS NMR Spectroscopy Study
* These authors contributed equally
In This Article
Summary
The motion regimes of ibuprofen encapsulated in β-cyclodextrin nanosponges polymer network are investigated using pulsed-field-gradient spin-echo (PGSE) NMR technique. Synthesis, purification, drug loading, implementation of the NMR pulse sequence and data analysis to work out the mean square displacement of the drug at several observation times are described in detail.
Abstract
The chemical cross-linking of β-cyclodextrin (β-CD) with ethylenediaminetetraacetic dianhydride (EDTA) led to branched polymers referred to as cyclodextrin nanosponges (CDNSEDTA). Two different preparations are described with 1:4 and 1:8 CD-EDTA molar ratios. The corresponding cross-linked polymers were contacted with 0.27 M aqueous solution of ibuprofen sodium salt (IP) leading to homogeneous, colorless, drug loaded hydrogels.
The systems were characterized by high resolution magic angle spinning (HR-MAS) NMR spectroscopy. Pulsed field gradient spin echo (PGSE) NMR spectroscopy was used to determine the mean square displacement (MSD) of IP inside the polymeric gel at different observation times td. The data were further processed in order to study the time dependence of MSD: MSD = f(td). The proposed methodology is useful to characterize the different diffusion regimes that, in principle, the solute may experience inside the hydrogel, namely normal or anomalous diffusion. The full protocols including the polymer preparation and purification, the obtainment of drug-loaded hydrogels, the NMR sample preparation, the measurement of MSD by HR-MAS NMR spectroscopy and the final data processing to achieve the time dependence of MSD are here reported and discussed. The presented experiments represent a paradigmatic case and the data are discussed in terms of innovative approach to the characterization of the transport properties of an encapsulated guest within a polymeric host of potential application for drug delivery.
Introduction
There is a growing interest in the design and formulation of polymeric systems capable of entrapping, via non-covalent interactions, small molecules with potential biochemical activity. Such materials are expected to find applications in the transport of the active principle to selective target and release upon the action of external stimuli, such as pH variations, temperature, etc. In this context, hydrogels turned out to be versatile and powerful materials for nanomedicine in view of controlled release of drugs1. The formation of polymeric hydrogels can be achieved by interconnecting the macromolecular chains by i) physical, non-covalent interactions such as hydrogen bonds, ii) covalent cross-linking of the chains leading to a three-dimensional network able to swell in the presence of an aqueous solution or iii) a combination of the two mentioned methods2-4.
A particularly versatile class of three-dimensional, swellable polymers for the encapsulation of organic and inorganic species can be obtained starting from natural β-cyclodextrin (β-CD) via condensation with suitable, activated derivatives of a tetracarboxylic acid5-8 giving rise to cyclodextrin nanosponges (CDNS). The synthesis, characterization and application of CDNS is a consolidated research theme of our group. The past few years' results indicate that CDNS show intriguing properties of swelling, absorption/inclusion of chemicals, and release of small drug molecules, with applications in controlled release of pharmaceutical active ingredients9-11 and environmental chemistry12-14.
Given these premises, two major issues to be addressed concern the efficient loading of the active compound in the polymeric gel and an improved understanding of solutes mobility in the gel matrices15. The literature provides both experimental studies and theories related to diffusion mechanisms of small molecules in macromolecular networks16,17. Pulsed field-gradient spin-echo (PGSE) NMR spectroscopy is a well-established structural method widely used to study the translational diffusion of small molecules in solvents18 or the self-diffusion of pure liquids. The recent developments of high resolution magic angle spinning (HR-MAS) NMR technology made it possible to collect high resolution NMR data of mobile molecules in heterogeneous suspensions19, gels and swellable polymers20,21. Indeed, the experimental setup combining HR-MAS NMR spectroscopy and the PGSE pulse sequence provides a unique opportunity to observe the solute molecules in the host's molecular environment. Important data on the transport properties of the entrapped drug molecule within a gel matrix can thus be obtained. High quality experimental data can thus be obtained allowing a more rational design of nanostructured host-guest systems.
In the present work we describe the detailed protocols for the following steps: i) synthesis and purification of two different formulation of CDNS cross-linked with EDTA polymers (Figure 1), referred to as CDNSEDTA, and characterized by different CD/cross linker molar ratio: 1:4 (CDNSEDTA 1:4) and 1:8 (CDNSEDTA 1:8); ii) the preparation of drug-loaded hydrogels for both CDNSEDTA 1:4 and CDNSEDTA 1:8. In this step we used, as model drug molecule, the popular non-steroidal anti-inflammatory ibuprofen sodium salt (IP); iii) the thorough investigation of the transport properties of IP within the CDNSEDTA hydrogels via PGSE-HRMAS NMR spectroscopy. The method we propose here is based on the measurement of the mean square displacement (MSD) of the encapsulated drug within the hydrogel followed by the analysis of the time dependence of the MSD.
We wish to stress that the methodology outlined above - which is focused on the time dependence of the drug's MDS in the matrix - provides a broader spectrum of information compared to the consolidated methodology based on the determination of the drug's diffusion coefficient only. We recently demonstrated21 that this approach allowed for the discrimination of normal and anomalous diffusion regimes experienced by IP confined in CDNS hydrogels.
We thus believe that the step-by-step description of polymer synthesis/purification, formation of the drug-loaded hydrogels, HR-MAS NMR characterization and data processing of MDS data, is a powerful toolkit for scientists interested in characterizing nanostructured systems for the confinement and release of small molecules.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Synthesis of CDNSEDTA Polymers
- Dry β-cyclodextrin (β-CD) in oven at 80 °C for 4 hr before use. Dry 500 ml of dimethylsulfoxide (DMSO) and 100 ml of triethylamine (Et3N) over molecular sieves (4 Å) for 24 hr before using them in the protocol.
- Introduce 25 ml of DMSO into a 50 ml one-neck round-bottom flask. Under magnetic stirring, add 5.675 g of β-CD (5 mmol). In order to reduce the formation of lumps, add the β-CD powder in small portions to DMSO.
- After about 30 min, add 6ml of Et3N to the homogeneous solution using a 10 ml graduated pipette. Keep the mixture under stirring for 15 min at RT. Plunge the flask into a water bath at RT.
NOTE: The reaction between β-CD and EDTA is exothermic. Therefore, plunging the flask into the water bath favors the heat exchange avoiding the overheating of the reaction mixture. - Add 5.124 g (20 mmol, preparation of CDNSEDTA 1:4) or 10.248 g (40 mmol, preparation of CDNSEDTA 1:8) of EDTA-dianhydride under intense stirring.
- After 3 hr, remove the solid material (CDNSEDTA 1:4 or CDNSEDTA 1:8) from the flask using a spatula and crush it grossly with a mortar and pestle.
- Wash the solid material onto filter paper with acetone at RT (100 ml × 5 times), with HCl 0.1 M (200 ml × 5 times), and deionized water (200 ml × 3 times).
- Finally, dry all the solid material in air at RT for 48 hr, crush it finely into a mortar and pestle and then keep it under vacuum (< 15 mbar) for 2 hr at 45 °C.
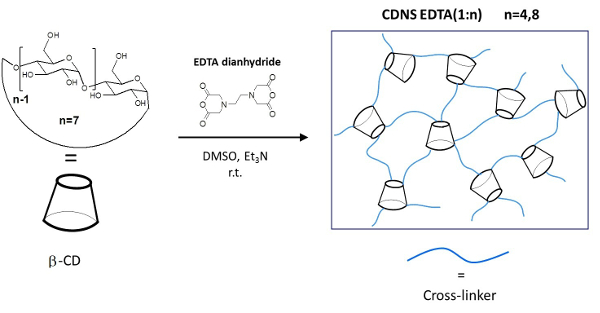
Figure 1: Schematic Representation of the CDNSEDTA Polymers. Schematic synthetic route. Left: molecular structure of the monomer β-cyclodextrin (β-CD) and cross-linking agent EDTA-dianhydride. On the arrow the overall reaction conditions. Right: sketch of the cross-linked polymer. Please click here to view a larger version of this figure.
2. HR-MAS NMR Measurements
- HR-MAS NMR Sample Preparation
- Prepare a solution 0.27 M of ibuprofen sodium salt (IP) in deuterated water (99.8%).
- Add 20 mg of CDNSEDTA 1:4 and 2 mg of anhydrous Sodium carbonate (Na2CO3) to 150 µl of the solution prepared at the point 2.1.1) into a 2 ml glass vial. Mix the content of the vial with a small spatula in order to homogenize it. Wait 2 hr before using the gel formed with this procedure. Repeat this point for the CDNSEDTA 1:8 polymer.
- Insert the gel in a 5mm NMR rotor suitable for HR-MAS NMR experiments using a small spatula. The total amount of gel to use depends on the internal volume of the rotor (12 μl recommended).
- HR-MAS 1H NMR Experiments
- Set the following instrumental parameters: rotor spinning speed of 4 KHz at the MAS pneumatic control unit, sample temperature at 305 K in the variable temperature unit.
- Acquire the 1H HR-MAS NMR spectra of ibuprofen in CDNSEDTA (1:4) and CDNSEDTA (1:8) polymer systems using a conventional one pulse sequence on the proton resonance.
- Create a new data set. Click on the "AcquPars" tab. Select the PULPROG: zg.
- Select the number of scans (NS = 4) and the time delay between them (D1 = 5 sec).Set the spectral width (SW = 8 ppm), the time domain (TD = 16K) and the receiver gain (RG = 32).
- Type "zg" on the console and there will be a free induction decay (FID) on the screen. To process the data click on the "ProcPars" tab. Set the spectral size (SI = 32K), an exponential multiplication window function (WDW = EM) and line broadening (LB = 1). Type "ft" to perform the Fourier transformation. Phase the spectrum using the phase tab on the screen. Obtain a high resolution well resolved spectrum.

Figure 2: The Bipolar Pulse Pairs Longitudinal Eddy Current Delay (BPPLED) Pulse Sequence. Schematic representation of the pulse sequence used to perform the PFGSE experiments. The phase cycle for the 90° pulses is: P1: (0)16, P2: (0022)4, P3: (0)4 (2)4 (1)4 (3)4, P4: (0202 2020 1313 3131), P5: (0)4 (2)4 (1)4 (3)4. The 180° pulses are + x. (modified from ref.18) Please click here to view a larger version of this figure.
- HR-MAS 1H NMR PGSE Experiments
NOTE: The PGSE experiments are performed using the BPPLED pulse sequence18 reported in Figure 2. This is a pseudo two-dimensional experiment with a gradient ramp increasing linearly from 2% to 100% in the indirect dimension. The signal intensity is attenuated depending on the diffusion time Δ and the gradient pulse δ. The optimization of these parameters is required before running properly a PGSE experiment. The optimization is done by running a few 1D measurements in which Δ is kept constant, while δ is varied.- Parameters Optimization
- Create a new data set - experiment number 1. Click on the "AcquPars" tab. Select the PULPROG: ledbpgp2s1d the 1D pulse sequence for diffusion optimization.
- Select the number of scans (NS = 16) and the time delay between them (D1 = 10 sec). Set the spectral width (SW = 8 ppm), the time domain (TD = 16K) and the receiver gain (RG = 32).
- Set Δ (D20 in the sequence) equal to a constant value and δ (p30) to a trial value. Start value Δ = 50 msec, δ = 3 msec (maximum allowed value for high resolution instruments).
- Read the value of spectral frequency (SFO1) from the 1H experiment and use now this value. Set the GPZ6 gradient strength to 2%. Repeat step 2.2.2.3. Use this spectrum as reference for the optimization.
- In the same data set create the experiment number 2. Observe all the experimental parameters. Increase the GPZ6 gradient strength to 95%. Repeat step 2.2.2.3. Compare this spectrum with the reference spectrum using the dual display icon and observe the change in the signal intensity.
NOTE: A well attenuated spectrum should have about 5% residual signal intensity compared with the reference spectrum. If the signal intensity is lost, reduce the value of δ and restart the section 2.3.1 procedure from point 2.3.1.3 until the right value for δ is found. - Repeat the parameters optimization procedure in section 2.3.1 for all the five Δ values.
NOTE: Choose five value for Δ = 50, 80, 110, 140 and 170 msec and optimized the corresponding δ to 3, 2.7, 2.4, 2.1, 1.8 msec (for IP in CDNSEDTA 1:8) and to δ to 2.7, 2.4, 2, 1.7, 1.4 (for IP in in CDNSEDTA 1:4).
- Acquisition of the 2D Diffusion Data Set
- In the same data set create the experiment number 3, all the 1D experimental parameters will be loaded. Type "eda". Select the PULPROG: ledbpgp2s the 2D pulse sequence and change the parmode to 2D.
- Set FnMODE=QF. Set the time domain TD in F2 dimension equal to 32, the number of gradient steps. All the other parameters are set correctly. Type "DOSY" and the gradient ramp will be generated and stored in a file. The start and final values of the ramp (2 - 95) are given as input parameters. The acquisition is now started.
- Parameters Optimization
- Data Processing
- Type "xf2" to execute the Fourier transformation in the F2 dimension. Type "abs2" to perform the baseline correction in the F2 dimension. Type "setdiffparm" to recall the experimental parameters (Δ, δ, and gradient list) for the next processing step.
- Click "T1/T2 relaxation module" in the analysis tab and define the peaks to be fitted using the first spectrum of the 2D experiment. Define the peak ranges and execute the fitting. The signal intensities at each applied gradient step are obtained.
NOTE: The signal intensities I(q, td), for each Δ value, depends on the experimental variables: applied pulse filed gradient (g), time variable (δ), magnetogyric ratio (γ) q= (γgδ) according to the following equation:

with the molecular MSD=z2. - Export the signal intensities in a spreadsheet and perform a linear fit of the data to get the value of z2 for each observed diffusion time td.
NOTE: The MSD value is related to the observation time td according to:
- Perform the log-log plot of z2 versus td for each experimental td value. The exponent α value is the slope of the linear regression. A more exhaustive discussion of the physical aspects of the equations reported above can be found in ref. 21 and in the references therein.
NOTE: Depending on the value of the exponent α, the diffusion regime is defined as: i) isotropic unrestricted diffusion for α = 1, ii) anomalous subdiffusive regime for 0 < α < 1, iii) anomalous superdiffusive regime for α > 1.
Access restricted. Please log in or start a trial to view this content.
Results
We first applied this methodology to the IP drug molecule dissolved in water solution in order to verify the viability of this approach. A full description of the representative results can be found in ref. 21. Rather, we will focus here on the methodological aspects and the nuts-and-bolts approach to data collection and data analysis. Figure 3 shows, on a semi-logarithmic scale, the normalized experimental signal decays I(q, td)/I(0, td) as a functi...
Access restricted. Please log in or start a trial to view this content.
Discussion
We present an experimental methodology to determine the diffusion regime of a small drug molecule encapsulated inside two representative formulations of CDNSEDTA hydrogels. HR-MAS PGSE NMR allows the determination of the mean square displacement of small molecules in a given diffusion time (in the range of a few milliseconds up to second), then monitoring distances in the micrometer scales. In the range observed (50 - 170 msec) only one type of motion is observed for each studied system. It should be stressed, however, t...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors gratefully acknowledge PRIN 2010-2011 NANOMED prot. 2010 FPTBSH and PRIN 2010-2011 PROxy prot. 2010PFLRJR_005 for funding.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| HR-MAS probe | BRUKER | N/A | Probe for NMR measurements on semi-solid samples |
| NMR Spectrometer | BRUKER | DRX 500 | FT NMR spectrometer for liquid ans semi-solis state |
| β-cyclodextrin (β-CD) | Alfa-Aesar | J63161 | Reagent |
| Ethylenediaminetetracetic (EDTA) dianhydride | Sigma-Aldrich | 332046 | Reagent |
| Dimethylsulfoxide (DMSO) | Alfa-Aesar | D0798 | Solvent |
| Triethylamine | Sigma-Aldrich | 471283 | Base (reagent) |
| Ibuprofen (IP) sadium salt | Sigma-Aldrich | I1892 | Antinflammatory drug |
| Excel 2010 | Microsoft | N/A | speadsheet for data analysis |
| Origin 8 SR0 | OriginLab Co. | speadsheet for data analysis |
References
- Sharpe, L. A., Daily, A. M., Horava, S. D., Peppas, N. A. Therapeutic applications of hydrogels in oral drug delivery. Expert Opin. Drug Deliv. 11, 901-915 (2014).
- Hennik, W. E., van Nostrum, C. F. Novel crosslinking methods to design hydrogels. Adv. Drug Deliv. Rev. 54, 13-36 (2002).
- Yu, L., Ding, J. D. Injectable hydrogels as unique biomedical materials. Chem. Soc. Rev. 37, 1473-1481 (2008).
- Ma, M., Kuang, Y., Gao, Y., Zhang, Y., Gao, P., Xu, B. Aromatic-Aromatic Interactions Induce the Self-Assembly of Pentapeptidic Derivatives in Water To Form Nanofibers and Supramolecular Hydrogels. J. Am. Chem. Soc. 132, 2719-2728 (2010).
- Trotta, F., Tumiatti, W. Patent WO. , 03/085002 (2003).
- Trotta, F., Tumiatti, W., Cavalli, R., Zerbinati, O., Roggero, C. M., Vallero, R. Ultrasound-assisted synthesis of cyclodextrinbased nanosponges. Patent WO. , 06/002814 (2006).
- Trotta, F., Cavalli, R. Characterization and applications of new hyper-cross-linked cyclodextrins. Compos. Interface. 16, 39-48 (2009).
- Cavalli, R., Trotta, F., Tumiatti, W. Cyclodextrin-based nanosponges for drug delivery. J. Incl. Phenom. Macrocycl. Chem. 56, 209-213 (2006).
- Trotta, F., et al. Cyclodextrin-based nanosponges as a vehicle for antitumoral drugs. Patent WO. , 09/003656 (2009).
- Vyas, A., Shailendra, S., Swarnlata, S. Cyclodextrin based novel drug delivery systems. J. Incl. Phenom. Macrocycl. Chem. 62, 23-42 (2008).
- Swaminathan, S., Vavia, P. R., Trotta, F., Torne, S. Formulation of beta-cyclodextrin based nanosponges of itraconazole. J. Incl. Phenom. Macrocycl. Chem. 57, 89-94 (2007).
- Mamba, B. B., Krause, R. W., Malefetse, T. J., Gericke, G., Sithole, S. P. Cyclodextrin nanosponges in the removal of organic matter to produce water for power generation. Water SA. 34, 657-660 (2008).
- Mamba, B. B., Krause, R. W., Malefetse, T. J., Nxumalo, E. N. Monofunctionalized cyclodextrin polymers for the removal of organic pollutants from water. Environ.Chem. Lett. 5, 79-84 (2007).
- Mhlanga, S. D., Mamba, B. B., Krause, R. W., Malefetse, T. J. Removal of organic contaminants from water using nanosponge cyclodextrin polyurethanes. J. Chem. Technol. Biot. 82, 382-388 (2007).
- Lehmann, S., Seiffert, S., Richtering, W. Spatially Resolved Tracer Diffusion in Complex Responsive Hydrogels. J. Am. Chem. Soc. 134, 15963-15969 (2012).
- Ferrer, G. G., Pradas, M. M., Ribelles, J. L. G., Colomer, F. R., Castilla-Cortazar, I., Vidaurre, A. Influence of the nature of the porous confining network on the sorption, diffusion and mechanical properties of hydrogel IPNs. Eur. Polym. J. 46, 774-782 (2010).
- Santoro, M., Marchetti, P., Rossi, F., Perale, G., Castiglione, F., Mele, A., Masi, M. Smart approach to evaluate drug diffusivity in injectable agar-carbomer hydrogels for drug delivery. J. Phys. Chem B. 115, 2503-2510 (2011).
- Johnson, C. S. Diffusion ordered nuclear magnetic resonance spectroscopy: principles and applications. Prog. Nucl. Magn. Reson. Spectroscopy. 34, 203-256 (1999).
- Viel, S., Ziarelli, F., Caldarelli, S. Enhanced diffusion-edited NMR spectroscopy of mixtures using chromatographic stationary phases. Proc. Natl. Acad. Sci. U. S. A. 100, 9696-9698 (2003).
- Alam, T. M., Hibbs, R. M. Characterization of heterogeneous solvent diffusion environments in anion exchange membranes. Macromolecules. 47, 1073-1084 (2014).
- Ferro, M., Castiglione, F., Punta, C., Melone, L., Panzeri, W., Rossi, B., Trotta, F., Mele, A. Anomalous diffusion of Ibuprofen in cyclodextrin nanosponges hydrogels: an HR-MAS NMR study. Beilstein J. Org. Chem. 10, 2715-2723 (2014).
- Wolf, G., Kleinpeter, E. Pulsed Field Gradient NMR Study of Anomalous Diffusion in a Lecithin-Based Microemulsion. Langmuir. 21, 6742-6752 (2005).
- Rossi, F., Castiglione, F., Ferro, M., Marchini, P., Mauri, E., Moioli, M., Mele, A., Masi, M. Drug-Polymer interactions in hydrogel-based drug-delivery systems: an experimental and theoretical study. Chem. Phys. Chem. , (2015).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved