A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Procedure for Adaptive Laboratory Evolution of Microorganisms Using a Chemostat
* These authors contributed equally
In This Article
Summary
Here, we present a protocol to obtain adaptive laboratory evolution of microorganisms under conditions using chemostat culture. Also, genomic analysis of the evolved strain is discussed.
Abstract
Natural evolution involves genetic diversity such as environmental change and a selection between small populations. Adaptive laboratory evolution (ALE) refers to the experimental situation in which evolution is observed using living organisms under controlled conditions and stressors; organisms are thereby artificially forced to make evolutionary changes. Microorganisms are subject to a variety of stressors in the environment and are capable of regulating certain stress-inducible proteins to increase their chances of survival. Naturally occurring spontaneous mutations bring about changes in a microorganism's genome that affect its chances of survival. Long-term exposure to chemostat culture provokes an accumulation of spontaneous mutations and renders the most adaptable strain dominant. Compared to the colony transfer and serial transfer methods, chemostat culture entails the highest number of cell divisions and, therefore, the highest number of diverse populations. Although chemostat culture for ALE requires more complicated culture devices, it is less labor intensive once the operation begins. Comparative genomic and transcriptome analyses of the adapted strain provide evolutionary clues as to how the stressors contribute to mutations that overcome the stress. The goal of the current paper is to bring about accelerated evolution of microorganisms under controlled laboratory conditions.
Introduction
Microorganisms can survive and adapt to diverse environments. Under severe stress, adaptation can occur via acquisition of beneficial phenotypes by random genomic mutations and subsequent positive selection1-3. Therefore, microbial cells can adapt by changing metabolic or regulatory networks for optimal growth, which is termed "adaptive evolution". Recent important microbial tendencies, such as outbreaks of superbugs and the occurrence of robust microbial strains, are very closely related to adaptive evolution under stressful conditions. Under defined laboratory conditions, we are able to study the mechanisms of molecular evolution and even control the direction of microbial evolution for various applications. Unlike multicellular organisms, single-celled organisms are well suited to adaptive laboratory evolution (ALE) for the following reasons: they regenerate quickly, they maintain large populations, and it is easy to create and maintain homogeneous environments. Combined with recent advances in DNA sequencing techniques and high-throughput technologies, ALE allows for the direct observation of genomic changes that lead to systemic regulatory changes. Mutational dynamics and a diversity of the population are also observable. Genetic engineering strategies can be determined from the analysis of ALE strains4,5.
Chemostat culture is a method used to obtain steady-state cells and increase productivity in fermentation processes6. Fresh medium is added and culture broth is harvested during the process (the latter includes medium and biomass). Long-term chemostat culture, however, changes the steady-state productivity of the culture and brings about the accumulation of spontaneous mutations and selection during culture (Figure 1a). Under various selection pressures (stressors), the accumulation of mutations is enhanced. A gradual increase of stress in a long-term chemostat provides for a continuous selection of mutations that work against the given stressors, such as temperature, pH, osmotic pressure, nutrient starvation, oxidation, toxic end products, etc. Colony transfer from a solid medium and serial transfer from a liquid medium (repeated batch culture) also allow researchers to obtain evolved microorganisms (Figure 1b and 1c). Although chemostat culture requires complicated methods, the pool of diversity (number of replications and population size) is higher than that obtained by colony transfer and serial transfer techniques. The stable stress exposure to individual cells and decreased variation in the cellular state during chemostat culture (steady state) are other benefits of ALE compared to batch culture-based techniques. Stress-induced ALE of Escherichia coli subjected to high succinate conditions is introduced in this article.

Figure 1: Methods of adaptive laboratory evolution. (A) Chemostat; (B) serial transfer; (C) colony transfer. The top figures illustrate the concept of the methods for ALE, and the bottom figures illustrate the number of cells that grew during ALE. Please click here to view a larger version of this figure.
Protocol
1. Equipment Preparation
- Obtain a chemostat jar (150-250 ml) or an Erlenmeyer flask (250 ml) containing an inlet port and an outlet port. Connect the ports with silicon tubing allowing for flow rates of 10-100 ml/hr. Optionally, use an air vent, an air outlet port, and temperature-controlled water inlet and outlet ports.
- Obtain a device suitable for the chemostat jar that provides for agitation and temperature control (or use a rotary shaking incubator).
- Obtain two peristaltic pumps in order to deliver fresh medium and collect the culture.
- Obtain a reservoir jar (10-20 L) containing a medium outlet port and an air inlet port.
- Obtain silicon tubing suitable for the dilution rate (i.e., ID 0.8 mm, flow range 0.06-36 ml/min; L/S 13 tubing).
2. Medium Preparation and Sterilization
- Initial Medium
- Dissolve 0.3 g glucose, 0.08 g NH4Cl, 0.05 g NaCl, 0.75 g Na2HPO4·2H2O, and 0.3 g KH2PO4 in 90 ml distilled water (D.W.) in a chemostat jar.
- Seal the chemostat jar along with the tubing using clamps. Do not seal the air vent.
- Sterilize the chemostat jar in an autoclave at 121 °C for 15 min. After sterilization, store the chemostat jar at room temperature.
- Dissolve 0.02 g MgSO4·7H2O, 0.01 g CaCl2, and 0.1 mg thiamine in 10 ml D.W. (solution A).
- Filter solution A using a syringe and a pre-sterilized syringe filter (a 0.45 µm pore filter).
- Add the solution A filtrates to the chemostat jar.
- Stress Medium
- Dissolve 30 g glucose, 8 g NH4Cl, 5 g NaCl, 75 g Na2HPO4·2H2O, 30 g KH2PO4, and 300 g disodium succinate hexahydrate (Na2·succinate·6H2O; the stressor used in this experiment) in 9.9 L D.W. in a reservoir jar.
- Seal the reservoir jar along with the tubing using clamps. Do not seal the air vent.
- Sterilize the reservoir jar in an autoclave at 121 °C for 15 min. After sterilization, store the jar at room temperature.
- Dissolve 2 g MgSO4·7H2O, 1 g CaCl2, and 10 mg thiamine in 100 ml D.W. (solution A).
- Filter solution A with a syringe and a pre-sterilized syringe filter (a 0.45 µm pore filter).
- Add the solution A filtrates to the reservoir jar.
- Aseptically connect the sterilized silicon tubing to the reservoir jar and attach the peristaltic pumps.
- High-stress Medium
- Prepare the medium as in section 2.2, but with a greater concentration of stressor (i.e., 3-5 g/L higher in the succinate adaptation).
Note: This protocol is for adaptation to a stress that can be delivered via the medium. In the case of physical stressors such as temperature, agitation, or illumination, the cultivation should be designed accordingly.
- Prepare the medium as in section 2.2, but with a greater concentration of stressor (i.e., 3-5 g/L higher in the succinate adaptation).
3. Initial Cultivation
- Inoculate a single colony of wild-type E. coli in a 15 ml test tube containing 4 ml of initial medium.
- Incubate the test tube in a shaking incubator for 12 hr at 37 °C and 220 rpm.
- Aseptically transfer 1 ml of preculture to the chemostat jar.
- Incubate the chemostat jar, providing for aeration (air 50 ml/min) and agitation (200 rpm), at 37 °C for 6 hr.
4. Stress Adaptation
- Aseptically connect the end of the silicon tubing from the pumps to the chemostat jar.
- Start the outlet pump (10 ml/hr or higher) and collect culture.
Note: The culture should be in the exponential phase, typically 4-8 hr after initial cultivation. - Check the optical density (600 nm) of the culture from the outlet tubing.
- Start the inlet pump (10 ml/hr, corresponding to a rate of dilution of 0.1 hr-1).
- Check the optical density of the culture at 600 nm from the outlet tubing every 24 hr.
- Operate the chemostat for 96 hr (9.6-fold turnover) or more. If the optical density is stable, exchange the reservoir containing the high-stress medium. If the optical density is lower than 0.2, stop the feeding inlet pump for 6 hr. Restart the inlet pump and check that the optical density is over 0.2.
- Gradually increase the concentration of the stressor by changing to a reservoir containing a higher stressor concentration.
- Take samples of the adapted culture whenever it reaches a milestone (e.g., a strain adapted to 100 g/L succinate stress), and store for further genomic analysis.
- For sample storage, mix the culture sample (0.5 ml) with a sterilized 80% glycerol solution (0.5 ml) and store it at -80 °C.
Note: If the microorganism acquires an ability to degrade the stressor during the ALE process, the stressor concentration in the fermentation jar is not the same as that in the fresh reservoir.
5. Single-colony Isolation of the Stress-adapted Strain
- Prepare agar plate medium (1.6% agar) containing the same stressor and at the same concentration of medium.
- Plate the outlet culture (0.1 ml) from the chemostat, and incubate at 37 °C for 16 hr.
- Pick single colonies from the plate using a sterile toothpick and inoculate them in 15 ml test tubes containing the same stressor and at the same medium concentration as in the chemostat, and incubate for 6 hr.
- Transfer 1 ml of culture broth into a 250 ml Erlenmeyer flask containing 50 ml of medium. Harvest 0.5 ml of the culture broth every 1 hr, and measure the O.D. at 600 nm. Compare the growth rate of the adapted strain to that of the wild-type strain given the stressor.
Results
For high-succinate stress adaptation, the wild-type E. coli W3110 strain was cultured in a chemostat at D = 0.1 hr-1 for 270 days (Figure 2).
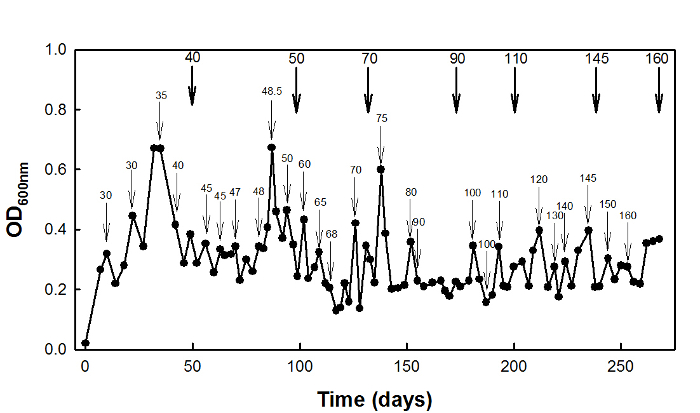
Figure 2: High-succinate stress adaptation of E. coli W3110 using chemostat culture. Thin arrows indicate the times at which the concentration of stressor was increased, and bold arrows i...
Discussion
Microorganisms are capable of adapting to almost all environments because of their rapid growth rate and genetic diversity. Adaptive laboratory evolution enables microorganisms to evolve under designed conditions, which provides a way of selecting individual organisms harboring spontaneous mutations that are beneficial under the given conditions.
The chemostat technique is more robust for achieving artificially driven evolution than transfer techniques for the following reasons: (a) a steady e...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study was financially supported by the Korean Ministry of Science, ICT and Future Planning (Intelligent Synthetic Biology Center program 2012M3A6A8054887). P. Kim was supported by a fellowship from the Catholic University of Korea (2015).
Materials
| Name | Company | Catalog Number | Comments |
| Mini-chemostat fermentor | Biotron Inc. | - | manufactured by special order |
| silicon tubing | Cole-Parmer | Masterflex L/S 13 | tubing size can be varied depending on the dilution rate and the size of fermentor jar. |
| reservoir jar | Bellco | Media storage bottle | 20 L |
| chemicals | Sigma-Aldrich | - | reagent grade |
| glucose | Sigma-Aldrich | G5767 | ACS reagent |
| NH4Cl | Sigma-Aldrich | A9434 | for molecular biology, suitable for cell culture, ≥99.5% |
| NaCl | Sigma-Aldrich | 746398 | ACS reagent, ≥99% |
| Na2HPO4·2H2O | Sigma-Aldrich | 4272 | 98.5-101% |
| KH2PO4 | Sigma-Aldrich | 795488 | ACS reagent, ≥99% |
| MgSO4·7H2O | Sigma-Aldrich | 230391 | ACS reagent, ≥98% |
| CaCl2 | Sigma-Aldrich | 793639 | ACS reagent, ≥96% |
| thiamine·HCl | Sigma-Aldrich | T4625 | reagent grade, ≥99% |
| Na2·succinate·6H2O | Sigma-Aldrich | S2378 | ReagentPlus, ≥99% |
References
- Rando, O. J., Verstrepen, K. J. Timescales of genetic and epigenetic inheritance. Cell. 128, 655-668 (2007).
- Kim, H. J., et al. Short-term differential adaptation to anaerobic stress via genomic mutations by Escherichia coli strains K-12 and B lacking alcohol dehydrogenase. Front Microbiol. 5, 476 (2014).
- Mendizabal, I., Keller, T. E., Zeng, J., Yi, S. V. Epigenetics and evolution. Integr Comp Biol. 54, 31-42 (2014).
- Lee, J. Y., Seo, J., Kim, E. S., Lee, H. S., Kim, P. Adaptive evolution of Corynebacterium glutamicum resistant to oxidative stress and its global gene expression profiling. Biotechnol Lett. 35, 709-717 (2013).
- Lee, J. Y., et al. Artificial oxidative stress-tolerant Corynebacterium glutamicum. AMB Express. 4, 15 (2014).
- Narang, A. The steady states of microbial growth on mixtures of substitutable substrates in a chemostat. J Theor Biol. 190, 241-261 (1998).
- Kwon, Y. D., Kim, S., Lee, S. Y., Kim, P. Long-term continuous adaptation of Escherichia coli to high succinate stress and transcriptome analysis of the tolerant strain. J Biosci Bioeng. 111, 26-30 (2011).
- Barrick, J. E., Lenski, R. E. Genome dynamics during experimental evolution. Nat Rev Genet. 14, 827-839 (2013).
- Li, H., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25, 2078-2079 (2009).
- McKenna, A., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297-1303 (2010).
- Deatherage, D. E., Barrick, J. E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 1151, 165-188 (2014).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved