A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Swab Sampling Method for the Detection of Human Norovirus on Surfaces
In This Article
Summary
A macrofoam based sampling methodology was developed and evaluated for the detection and quantification of norovirus on environmental hard surfaces.
Abstract
Human noroviruses are a leading cause of epidemic and sporadic gastroenteritis worldwide. Because most infections are either spread directly via the person-to-person route or indirectly through environmental surfaces or food, contaminated fomites and inanimate surfaces are important vehicles for the spread of the virus during norovirus outbreaks.
We developed and evaluated a protocol using macrofoam swabs for the detection and typing of human noroviruses from hard surfaces. Compared with fiber-tipped swabs or antistatic wipes, macrofoam swabs allow virus recovery (range 1.2-33.6%) from toilet seat surfaces of up to 700 cm2. The protocol includes steps for the extraction of the virus from the swabs and further concentration of the viral RNA using spin columns. In total, 127 (58.5%) of 217 swab samples that had been collected from surfaces in cruise ships and long-term care facilities where norovirus gastroenteritis had been reported tested positive for GII norovirus by RT-qPCR. Of these 29 (22.8%) could be successfully genotyped. In conclusion, detection of norovirus on environmental surfaces using the protocol we developed may assist in determining the level of environmental contamination during outbreaks as well as detection of virus when clinical samples are not available; it may also facilitate monitoring of effectiveness of remediation strategies.
Introduction
Human noroviruses are a leading cause of epidemic and sporadic acute gastroenteritis worldwide 1,2,3. The virus is extremely contagious and transmission occurs through direct person to person interaction or indirectly through contact with contaminated food, water or environmental surfaces. Noroviruses can be shed for extended periods and prolonged survival of the virus on environmental surfaces has been documented 1,2,3. During peak shedding, billions of virus particles are released per gram of feces, and vomit also contains a sufficient number of viral particles to cause infection 4,5,6,7,8,9,10. In addition, transfer of the virus between inanimate surfaces and human skin can occur easily 2,11,12. Hence, monitoring of environmental contamination may assist in outbreak investigations and in assessing the effectiveness of clean-up and disinfection procedures.
Several environmental sampling protocols have been described for the detection of rotavirus, coliphage MS2, feline calicivirus (FCV), and bacteriophage P22 13,14,15,16. However, the validation conditions described in these studies, including fast desiccation (<1 hr) and small surface areas (25 x 100 cm2), may not adequately represent field settings. In addition, the expected low contamination levels of environmental surfaces require protocols that are able to detect very few virus particles.
We developed a macrofoam-based surface sampling method for the detection and typing of norovirus. This method has been validated during several norovirus outbreaks. The protocol includes 1) how to collect swab samples from environmental surfaces (2) how to best maintain integrity of the samples during collection and shipping to the laboratory, and 3) laboratory testing and typing of norovirus.
Protocol
1. Swab Sampling in the Field
- Wear a clean pair of gloves.
- Measure the size of the sampling area without touching the surface using a measuring tape or ruler. Try to estimate the area as accurately as possible and fill out a report form (Supplementary Table 1).
- Check the swab kit for possible leakages and label sample transport bags and swab kits.
- Move the swab across the sampling area as follows: one stroke in horizontal direction, one stroke in vertical direction, and one stroke in a diagonal direction. Do not swab a surface area larger than 700 cm2.
- Place each swab into a tube and tighten the cap securely.
2. Storage and Transport of Swabs to the Laboratory
- Keep swabs at 4 °C for up to 48 h. If storage for longer periods is required, store the swabs at -20 °C (or -70 °C).
- Keep the tubes at 0-4 °C (i.e., use cold packs) in an insulated container during transport to the laboratory and ship within 24-48 h of collection.
3. Virus Concentration, Viral RNA Extraction and Purification
NOTE: All centrifugation steps use a table top centrifuge at 5,000 x g for 5 min at room temperature, unless stated otherwise. Be extra careful when working with the universal nucleic acid extraction (UNEX) buffer. Wear goggles or face shield.
- Label one 15 mL tube and one RNA Midi column for each sample. Include a negative extraction control in each experiment.
- To prepare the Lysis solution for 10 swabs, mix 25 ml of UNEX buffer with 25 mL of PBST (PBS pH 7.2 containing 0.02% Tween-80).
- Add 50 µL of coliphage MS2 suspension (106 PFU/µl) to 50 mL of Lysis solution.
- Place a swab in a 15 mL tube and add 5 ml Lysis solution. Mix and incubate for 10 min at room temperature.
- Add 5 ml 100% ethanol to each tube and vortex for 10 s.
- Carefully remove the swab from the 15 mL tube (it contains UNEX buffer) pressing it gently against the side of the tube to remove excess liquid and then discard the swab. The remaining volumes should be between 9-10 mL.
4. Midi Column Viral Nucleic Acid Extraction
- Carefully transfer 4.5 mL UNEX/ethanol mixture from step 3.6 onto a midi column, centrifuge and discard the filtrate.
- Load another 4.5 mL from the same mixture onto the same column, centrifuge, and discard the filtrate.
- For the first wash, add 3.5 ml of 70% ethanol onto the Midi column, centrifuge and discard the filtrate.
- For the second wash, add another 3.5 ml of 70% ethanol onto the Midi column. Centrifuge and discard the filtrate.
- For the dry-spin, centrifuge the Midi columns to remove all traces of ethanol (which can negatively affect your viral RNA recovery).
- Place the Midi column in a new 15 ml centrifuge tube. Add 250 µl elution buffer onto the Midi column; wait for 1 min before centrifuging to obtain the highest viral RNA recovery.
- Store the extracted nucleic acid at -70 °C or proceed immediately to step 5.
5. Concentration of Viral Nucleic Acid Using RNA Clean and Concentrator Kits
- Add 500 µL of RNA Binding Buffer to 250 µL of RNA eluted in step 4.7 above and vortex for 10 s.
- Add 750 µL 100% ethanol and vortex for 10 s.
- Label a spin column for each sample. Load the 750 µL sample onto a spin column.
- Centrifuge the spin columns at 12,000 x g for 1 min. Discard the flow-through.
- Load the remaining 750 µL onto a spin column and centrifuge at 12,000 x g for 1 min.
- For the pre-wash, add 400 µL RNA Prep buffer to each spin column and centrifuge at 12,000 x g for 1 min. Discard the flow-through.
- For the first wash, add 800 µL RNA Wash Buffer to each spin column and centrifuge at 12,000 x g for 1 min. Discard the flow-through.
- For the second wash, add 400 µL RNA Wash Buffer to spin column and centrifuge at 12,000 x g for 1 min. Discard the flow-through.
- For the dry-spin, centrifuge the spin column to remove all traces of wash buffer at 12,000 x g for 2 min.
- Carefully transfer spin column to a clean 1.5 mL microcentrifuge tube.
- Add 25 µL elution buffer onto the spin column and incubate for 1 min at room temperature. This will increase the recovery of viral RNA from the column.
- Collect RNA by centrifuging the spin column at 10,000 x g for 1 min.
- Either proceed directly to RT-qPCR (step 6) or store RNA at -70 °C.
6. Mutiplex RT-qPCR Detection of Genogroup I, and II Noroviruses, and Coliphage MS 2 (Supplementary Table 2)
- Clean working surfaces, pipettes, and centrifuges with RNase decontamination solution to reduce possible contamination.
- Thaw RT-qPCR reagents on ice. Thaw RNA on ice (in a separate area/room).
- Determine the number of reactions and add at least 10% more of them (e.g. if 10 reactions are needed, make master mix for 11).
- Vortex individual master mix components, except for 25x RT-qPCR enzyme, for 5 s. Carefully mix the enzyme by flicking the tube with a finger.
- Briefly centrifuge master mix components for 5 s.
- Prepare the master mix for the realtime RT-PCR detection of norovirus GI and GII according to the kit instructions and add norovirus-specific oligonucleotide primers and probes in a 1.5 ml microcentrifuge tube (Supplementary Table 3).
- Mix the master mix by pipetting 5-10 times up and down (vortexing is not recommended). Aliquot 22 µL of the master mix in each well of real time PCR 96-well plate.
- Vortex sample RNA for 5 sec, and collect by brief (5 s) centrifugation.
- Add 3 µL of sample RNA and GI and GII positive controls to the RT-qPCR plate (follow template from Table 2). Add 3 µL of nuclease-freewater in the no-template-control (NTC)wells.
- Seal the real-time plate with optical adhesive film.
- Carefully centrifuge the real-time plate at 1,300 x g for 1 min to remove any air bubbles or liquid drops that may be present in the wells.
- Set up a real-time PCR instrument and set-up the following thermocycling conditions: 1) RT step for 10 min. at 45 °C (2) activation of Taq polymerase, 10 min. at 95 °C and (3) 45 cycles of 15 s at 95 °C, and 60 s at 60 °C.
7. Quantification of Norovirus in Swab Samples
- Check the results of positive and negative controls to validate RT-qPCR results (Supplementary Table 3).
- Determine whether each standard curve meets the acceptable values given in Supplementary Table 2 and calculate the overall standard deviation for the standard curve using Equation 1.
(Equation 1) % efficiency = 100 x 10(average Ct Values-Intercept)/Slope. - If the PCR thermal cycler software does not calculate the slope for each standard curve, determine slope and R2values by regression using statistic software (e.g. Excel, SPSS, or SAS). In addition, calculate the % efficiency for standard curves following equation I
- Record the RNA copy number calculated by the thermal cycler software for all test samples based upon standard curves that meet the criteria specified in Supplementary Table 4. Rerun any samples with standard curves that do not meet the criteria or have false-positive controls. Check Ct values of coliphage MS2, which was added as an internal control to monitor PCR inhibition.
- Determine the total number of RNA copies per sample by the multiplying the RNA copy number calculated in step 7.2 by the ratio of the volume (25 to 50 µL) of total RNA eluent to that of the RNA (3 to 5 µL) used for the RT-qPCR reaction. Alternatively, calculate the RNA density by dividing the total RNA copy number by the object surface area (cm2) analyzed.
8. Genotyping of Real-time RT-PCR Positive Samples by Hemi Nested Conventional PCR Amplification
- Prepare the first round of master mix for each primer set of the RT-PCR assay (Supplementary Tables 2 , and 5).
- Add 5 µL of norovirus positive RNA to 20 µL of master mix.
- Run the RT-PCR under the following conditions: (1) RT step for 30 min at 42 °C (2) activation of Taq polymerase, 15 min at 95 °C, and (3) 40 cycles of 30 s at 95 °C, 30 s at 50 °C, and 60 sec at 72 °C. After 40 cycles, incubate for an additional 10 min at 72 °C.
- Prepare the second round of mater mix for each primer set of the RT-PCR assay (Supplementary Tables 2 and 5).
- Add 23 µL master mix and add 2 µL of first-round RT-PCR products (from first round) to each tube (ideally the first-round product should be diluted 1/10 in RNase-free water).
- Repeat step 8.3.
- Prepare a 2% agarose gel in 100 ml 1x Tris-acetate EDTA (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA, at pH 8.3). After dissolving agarose by heating the mixture in a microwave oven, add 10 µL of nucleic acid stain per 100 mL of prepared gel after cooling the mixture to 60-70 °C, pour the gel and insert the combs. Let the gel settle for at least 30 min.
- Mix 15 µl of each RT-PCR product with 3 µL of 6x loading dye. Load 15 µL of each sample and electrophorese the agarose gel at 100 V for 1 h.
- Excise target RT-PCR fragments of appropriate size (330 bp for GI and 341 bp for GII) from the gel and purify RNA using a commercial gel extraction kit. The purified PCR product can now be used for Sanger sequencing.
Results
Figure 1 presents a flowchart of the swab sampling protocol. This protocol consists of four main steps; 1) sample collection, 2) sample storage and transportation, 3) viral RNA purification and concentration and 4) RT-qPCR assay and genotyping.
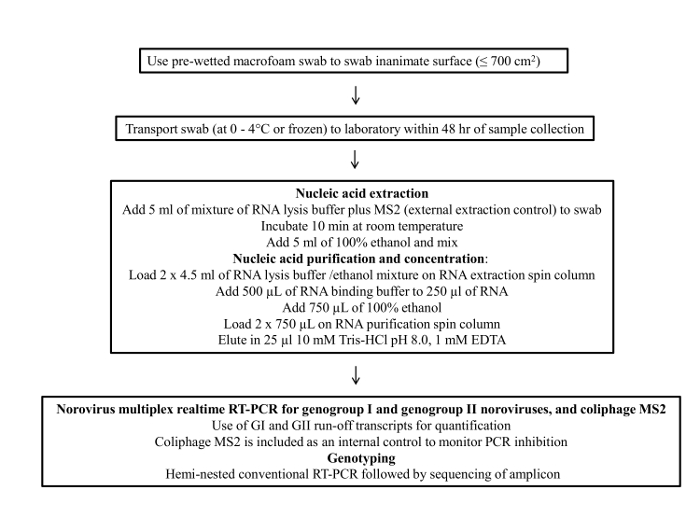
Figure 1: Flow chart of the final protocol for environmental surface sampling of noroviru...
Discussion
Noroviruses have a 50% human infectious dose between 18 and 103 virus particles20. Therefore, even low-level contamination of surfaces may pose a public health risk. Several aspects of the swab sampling protocol were evaluated including: 1) different swab materials, 2) storage condition swabs during transport, 3) viral RNA concentration, and 4) coliphage MS2 as internal extraction control.
Until recently, only the performance of swabs made from cotton, polyes...
Disclosures
Authors have no conflicting interest. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Acknowledgements
The authors have no acknowledgements.
Materials
| Name | Company | Catalog Number | Comments |
| Generic name for kits | |||
| Macrofoam swab | Premoistened EnviroMax Swab kit | Puritan | 2588060PFUW |
| RNA Lysis buffer | CDC UNEX buffer | Microbiologics | Cat No MR0501 |
| RNA extraction spin column | Midi column | Omega Biotek | Cat No R6664-02 |
| RNA purification spin column | Zymol RNA Clean and Concentrator kit | Zymo Research | Cat No R1016 |
| Real time RT-PCR kit | AgPath kit One-Step RT-PCR Kit | Life Technologies | Cat No 4387391 |
| Conventional RT-PCR kit | Qiagen one step RT-PCR kit | Qiagen kit | Cat No 210212 |
| Gel extraction kit | Qiagen QIAquick gel extraction kit | Qiagen kit | Cat No 28704 or 28706 |
| Coliphage MS2 | ATCC | Cat No 15597-B1 | |
| RNA run-off transcripts | |||
| Realtime PCR platform | Applied Biosystems | Model ABI 7500 | |
| Optical 96-well reaction plate | Thermo Scientific | Cat No 4316813 | |
| MicroAmp Clear Adhesive Film | Thermo Scientific | Cat No 4306311 |
References
- Isakbaeva, E. T., et al. Norovirus transmission on cruise ship. Emerg. Infect. Dis. 11, 154-158 (2005).
- Lopman, B. A., Gastañaduy, P., Park, G. W., Hall, A. J., Parashar, U. D., Vinjé, P. Environmental transmission of norovirus gastroenteritis. Curr. Opin. Virol. 2 (1), 1-7 (2011).
- Malek, M., et al. Outbreak of norovirus infection among river rafters associated with packaged delicatessen meat, Grand Canyon, 2005. Clin Infect Dis. 48 (1), 31-37 (2009).
- Atmar, R. L., et al. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 14 (10), 1553-1557 (2008).
- Glass, R. I., Parashar, U. D., Estes, M. K. Norovirus gastroenteritis. N. Engl. J. Med. 361 (18), 1776-1785 (2009).
- Park, G. W., et al. Evaluation of a New Environmental Sampling Protocol for Detection of Human Norovirus on Inanimate Surfaces. Appl. Environ. Microbiol. 81 (17), 5987-5992 (2015).
- Barker, J., Jones, M. V. The potential spread of infection caused by aerosol contamination of surfaces after flushing a domestic toilet. J. Appl. Microbiol. 99, 339-347 (2005).
- Tung-Thompson, G., Libera, D. A., Koch, K. L., de Los Reyes, F. L., Jaykus, L. A. Aerosolization of a Human Norovirus Surrogate, Bacteriophage MS2, during Simulated Vomiting. PloS one. 10, 0134277 (2015).
- Atmar, R. L., et al. Determination of the 50% human infectious dose for Norwalk virus. J. Infect. Dis. 209 (7), 1016-1022 (2014).
- Petrignani, M., van Beek, J., Borsboom, G., Richardus, J. H., Koopmans, M. Norovirus introduction routes into nursing homes and risk factors for spread: a systematic review and meta-analysis of observational studies. J. Hosp. Infect. 89 (3), 163-178 (2015).
- . Centers for Disease Control Prevention. Norovirus outbreak in an elementary school--District of Columbia, February 2007. MMWR. Morb. Mortal. Wkly. Rep. 56 (51-52), 1340-1343 (2008).
- Cheesbrough, J. S., Barkess-Jones, L., Brown, D. W. Possible prolonged environmental survival of small round structured viruses. J. Hosp. Infect. 35, 325-326 (1997).
- Julian, T. R., Tamayo, F. J., Leckie, J. O., Boehm, A. B. Comparison of surface sampling methods for virus recovery from fomites. Appl. Environ. Microbiol. 77, 6918-6925 (2011).
- Taku, A., et al. Concentration and detection of caliciviruses from food contact surfaces. J. Food. Prot. 65, 999-1004 (2002).
- Scherer, K., Ellerbroek, L., Schulenburg, J., Johne, R., Klein, G. Application of a swab sampling method for the detection of norovirus and rotavirus on artifically contaminated food and environmental surfaces. Food. Environ. Virol. 1 (42), 42-49 (2009).
- Herzog, A. B., et al. Evaluation of sample recovery efficiency for bacteriophage P22 on fomites. Appl. Environ. Microbiol. 78, 7915-7922 (2012).
- Vega, E., et al. CaliciNet: A Novel Surveillance Network for Norovirus Gastroenteritis Outbreaks in the United States. Emerging Infectious Diseases. 17 (8), 1389-1395 (2011).
- Rolfe, K. J., et al. An internally controlled, one-step, real-time RT-PCR assay for norovirus detection and genogrouping. J Clin Virol. 39 (4), 318-321 (2007).
- Kittigul, L., et al. Norovirus GII-4 2006b variant circulating in patients with acute Thailand during a 2006-2007 study. J. Med. Virol. 82 (5), 854-860 (2010).
- Teunis, P. F., et al. Norwalk virus: how infectious is it. J. Med. Virol. 80 (8), 1468-1476 (2008).
- Wollants, E., et al. Evaluation of a norovirus sampling method using sodium dodecyl sulfate/EDTA-pretreated chromatography paper strips. J. Virol. Methods. 122, 45-48 (2004).
- Weir, M. H., Shibata, T., Masago, Y., Cologgi, D., Rose, J. B. The Effect of Surface Sampling and Recovery of Viruses and Non-Spore Forming Bacteria on a QMRA Model for Fomites. Environ. Sci. Technol. 50 (11), 5945-5952 (2016).
- . Microbiology of food and animal feed-Horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR. International Organization for Standardization (ISO). , (2013).
- Huslage, K., Rutala, W. A., Sickbert-Bennett, E., Weber, D. J. A quantitative approach to defining "high-touch" surfaces in hospitals. Infect. Control. Hosp. Epidemiol. 31 (8), 850-853 (2010).
- Wu, H. M., et al. A norovirus outbreak at a long-term-care facility: the role of environmental surface contamination. Infect. Control. Hosp. Epidemiol. 26 (10), 802-810 (2005).
- Ikner, L. A., Gerba, C. P., Bright, K. R. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 4 (2), 41-67 (2012).
- Gallimore, C. I., et al. Environmental monitoring for gastroenteric viruses in a pediatric primary immunodeficiency unit. J. Clin. Microbiol. 44 (2), 395-399 (2006).
- Ganime, A. C., et al. Dissemination of human adenoviruses and rotavirus species A on fomites of hospital pediatric units. Am J Infect Control. , (2016).
- Verani, M., Bigazzi, R., Carducci, A. Viral contamination of aerosol and surfaces through toilet use in health care and other settings. Am J Infect Control. 42 (7), 758-762 (2014).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved