A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
CRISPR-Cas9-based Genome Engineering to Generate Jurkat Reporter Models for HIV-1 Infection with Selected Proviral Integration Sites
In This Article
Summary
We present a genome engineering workflow for the generation of new in vitro models for HIV-1 infection that recapitulate proviral integration at selected genomic sites. Targeting of HIV-derived reporters is facilitated by CRISPR-Cas9-mediated, site-specific genome manipulation. Detailed protocols for single-cell clone generation, screening, and correct targeting verification are provided.
Abstract
Human immunodeficiency virus (HIV) integrates its proviral DNA non-randomly into the host cell genome at recurrent sites and genomic hotspots. Here we present a detailed protocol for the generation of novel in vitro models for HIV infection with chosen genomic integration sites using CRISPR-Cas9-based genome engineering technology. With this method, a reporter sequence of choice can be integrated into a targeted, chosen genomic locus, reflecting clinically relevant integration sites.
In the protocol, the design of an HIV-derived reporter and choosing of a target site and gRNA sequence are described. A targeting vector with homology arms is constructed and transfected into Jurkat T cells. The reporter sequence is targeted to the selected genomic site by homologous recombination facilitated by a Cas9-mediated double-strand break at the target site. Single-cell clones are generated and screened for targeting events by flow cytometry and PCR. Selected clones are then expanded, and correct targeting is verified by PCR, sequencing, and Southern blotting. Potential off-target events of CRISPR-Cas9-mediated genome engineering are analyzed.
By using this protocol, novel cell culture systems that model HIV infection at clinically relevant integration sites can be generated. Although the generation of single-cell clones and verification of correct reporter sequence integration is time-consuming, the resulting clonal lines are powerful tools to functionally analyze proviral integration site choice.
Introduction
Integration of proviral DNA into the host genome upon infection is a critical step in the life cycle of human immunodeficiency virus (HIV). Following integration, HIV persists by establishing latency in long-lived CD4+ T cell subsets such as memory CD4+ T cells. HIV integration appears to be non-random1,2. A number of genomic hotspots with recurrently integrated proviral DNA has been detected in several studies through the sequencing of integration sites in acutely and chronically infected individuals2,3,4,5,6,7,8. Interestingly, at some of these integration sites, the same locus was detected in a large fraction of infected cells, leading to the idea that integration at recurrent sites might positively affect clonal expansion1.
To advance our understanding of the significance of recurrent integration sites, proviral integration site choice must be explored. However, several technical aspects hamper studying HIV integration site choice and the consequences. Broadly used cell culture models for HIV latency like JLat cell lines do not reflect clinically relevant recurrent integration sites9. Studies on primary patient-derived cells, on the one hand, enable description of integration site landscape by sequencing but do not allow for functional analyses. To our knowledge, no adequate experimental model is available to functionally analyze selected clinically relevant integration sites.
Here we present a detailed workflow to generate novel models for HIV infection using CRISPR-Cas9-based genome engineering technology. The workflow described herein can be used to generate T cell-derived reporter cell lines that model HIV infection, carrying a genomically integrated proviral reporter at a chosen integration site. They are thus serving as new tools to explore how the proviral integration site can impact HIV biology and how the provirus responds to different treatment strategies (e.g., inducibility by latency reversing agents). Our method uses the advantages of CRISPR-Cas9-based genome engineering, in which integration of the reporter sequence by homologous recombination is facilitated by a Cas9 nuclease-induced double-strand break at the target site. Target sites for integration are chosen according to proximity to the described recurrent integration sites from studies on HIV-infected individuals and the presence of suitable PAM motifs for Cas9-mediated genome engineering.
In our exemplary results, we have focused on the BACH2 gene locus, which codes for the BTB And CNC Homology transcriptional regulator 2. In chronically HIV-infected individuals on antiretroviral therapy, BACH2 is one of the loci showing enrichment of integrated HIV-1 sequences3,6,7,8,10. We have chosen a minimal HIV-derived reporter consisting of HIV-1-derived long terminal repeat (LTR), tdTomato coding sequence, and bovine growth hormone (BGH) polyadenylation signal (PA), which we have targeted to two specific sites in BACH2 intron 5. The presented protocol is optimized for Jurkat cells, a human CD4+ T cell-derived suspension cell line, but other cell lines may be used and the protocol adapted accordingly. We present a detailed workflow for selection of target site, construction of target vector with homology arms, CRISPR-Cas9-mediated targeting of the reporter into the chosen genomic site, generation and selection of clonal lines, and comprehensive verification of newly generated, targeted reporter cell lines.
Protocol
1. Targeting Strategy for Genome Engineering and Targeting Vector (tv) Design
NOTE: The first step of genome engineering involves selection and generation of the necessary tools for CRISPR-Cas9-mediated targeting. Selection of a genomic integration site locus, choice of cell type for targeting, and design of an HIV-derived reporter for integration should precede this step. This protocol describes targeting of an HIV-LTR_tdTomato_BGH-PA minimal reporter into Jurkat target cells. A flow chart of the workflow for CRISPR-Cas9-based targeting, generation, screening and verification of clonal lines is depicted in Figure 1. The described targeting strategy uses the S. pyogenes Cas9 (SpCas9) to generate gRNA-directed dsDNA breaks at a selected integration site. The reporter is then targeted into the chosen genomic locus through homologous recombination by providing a non-linearized targeting vector (tv) that contains the reporter sequence flanked by so-called 5’ and 3’ homology arms (HA)11.
- Choice of targeted locus, gRNA, and targeting vector design
- Choose thegenomic locus to be targeted based on the individual scientific question. Use published lists of recurrent integration sites of HIV found in patients in different studies2,3,4,5,6,7,8 as a guideline. In silico extract the genomic sequence of the desired genomic locus to be targeted (sequence of the complete gene or at least 5 kb of genomic sequence) using UCSC genome browser (http:// genome.edu.ucsc.edu).
- Choose guide RNAs (gRNAs) of 20 nt for targeting of the chosen genomic locus using the E-CRISP webtool (http://www.e-crisp.org).
- Select “Homo sapiens GRCh38” as the organism. Input 2,000 bp of the genomic sequence covering the desired genomic locus extracted in step 1.1.1.
- Start a gRNA search using medium application settings (any PAM, any 5’ base, off-targets tolerate mismatches, and introns/CPG islands are excluded). A list with possible gRNA designs will appear, ranking from highest to lower scores for specificity and efficiency.
- Select a gRNA that preferably shows a high score for specificity and efficiency and is as close as possible to the desired genomic locus to be targeted.
NOTE: A compromise between proximity to the desired genomic locus and design of specific and efficient gRNA has to be found.
- Blast the chosen gRNA sequence against the reference genome using the NCBI blast browser (https://blast.ncbi.nlm.nih.gov) to check for uniqueness of the gRNA binding site.
- Select “human” as the genome. Input the gRNA sequence as the query sequence. Select “highly similar sequences” (megablast) as the program. Ensure that the gRNA sequence is unique. If not, chose a different gRNA from step 1.1.2.3 and blast again.
- Once gRNA sequence is chosen, select in silico 1,000 bp upstream and downstream of gRNA sequence from genomic sequence extracted in step 1.1.1 as 5’ and 3’ HA accordingly.
NOTE: The gRNAs should be homologous to the chosen genomic integration site locus and located adjacent to a protospacer adjacent motif (PAM; e.g., NGG for SpCas9) (Figure 2a). The tv contains the reporter sequence that is 5’ and 3’ flanked by HAs. HAs cover 1000 bp upstream and downstream of the gRNA sequence11. The full gRNA sequence should not be included in the HA. An overlap of up to 5 nt is acceptable (Figure 2a).
- Generation of gRNA and targeting vectors
NOTE: For vector schemes, see Figure 2b.- To generate a vector for expression of SpCas9 and gRNA, use the pX330-U6-Chimeric_BB-cBh-hSpCas9 as the backbone from which both SpCas9 and the single guide RNA (sgRNA) can be simultaneously expressed. To clone the gRNA sequence into the backbone, use the BbsI restriction sites12.
- To generate the tv, choose a high-copy plasmid as backbone (e.g., pMK or cDNA3.1).
- First, assemble the reporter (in this protocol: LTR_tdTomato_BGH-PA) into the construct backbone by Gibson Assembly clong13 using a commercial assembly cloning kit, and introduce 5’ and 3’ flanking restriction sites (e.g., 5’ PacI and 3’ SmaI) for subsequent restriction digestion cloning of the HAs.
- Amplify 1000 bp of the HA fragments chosen in step 1.1.4 from genomic DNA (gDNA) of the cell type to be targeted (in this protocol: Jurkat cells) using a DNA polymerase with proofreading activity (see Tables 1 and 2 for PCR ingredients and cycling conditions). Then, introduce reporter flanking restriction sites on the 5’ and 3’ ends of each HA (PacI on 5’ HA on both ends, SmaI on 3’ HA on both ends).
- Sequentially clone HAs into construct backbone already containing the reporter (generated in step 1.2.2.1) by restriction enzyme cloning14,15. First, clone in 5’ HA using PacI restriction sites, then clone in 3’ HA using SmaI restriction sites.
NOTE: If tv backbone contains an additional fluorescent reporter, unwanted backbone integration can be assessed by flow cytometry (see steps 3.2.2 and 3.2.3). If tv backbone contains no fluorescent reporter, backbone integration must be assessed using PCR (see step 3.2.8).
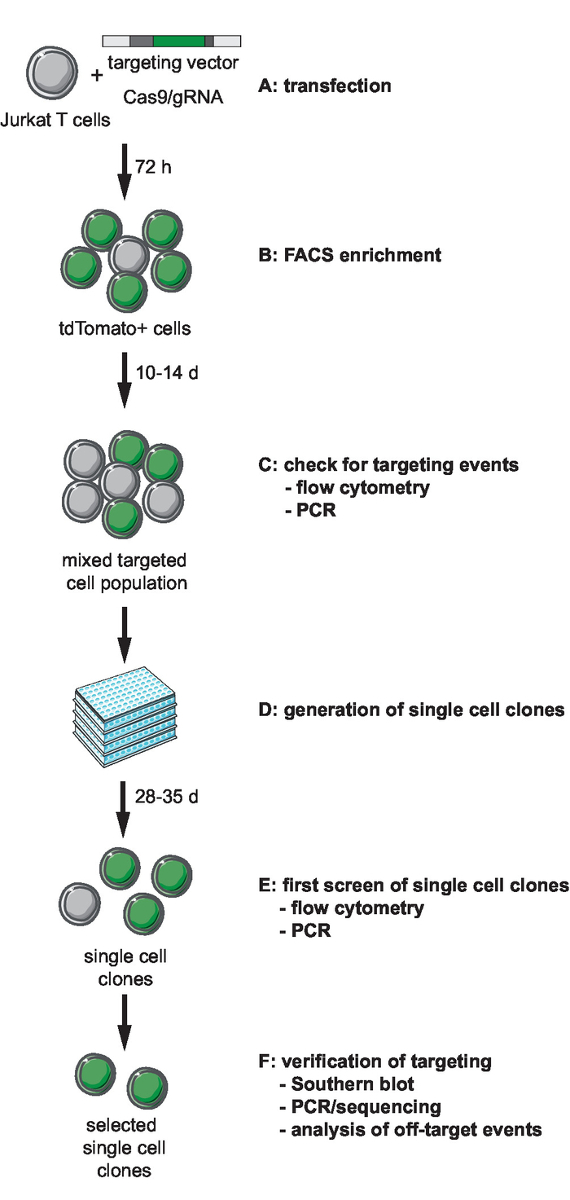
Figure 1: Workflow for CRISPR-Cas9-mediated targeting, generation, and selection of clonal reporter lines with defined integration site. (A) Generate the target vector and transduce Jurkat T cells with the target vector and Cas9/gRNA expression plasmid. (B) Enrich the transfected cells 72 h post transfection by FACS. (C) Let the cells grow for 10 to 14 days and confirm the occurrence of targeting events by PCR and flow cytometry. (D) Generate single-cell clones by limiting dilution and let clones grow for 3 weeks. (E) Screen the clones for correct targeting by PCR and flow cytometry in 96-well format. Expand selected clones. (F) Verify correct targeting in selected clones by Southern blot, PCR and sequencing, and analysis of off-target events of Cas9 endonuclease activity. Please click here to view a larger version of this figure.
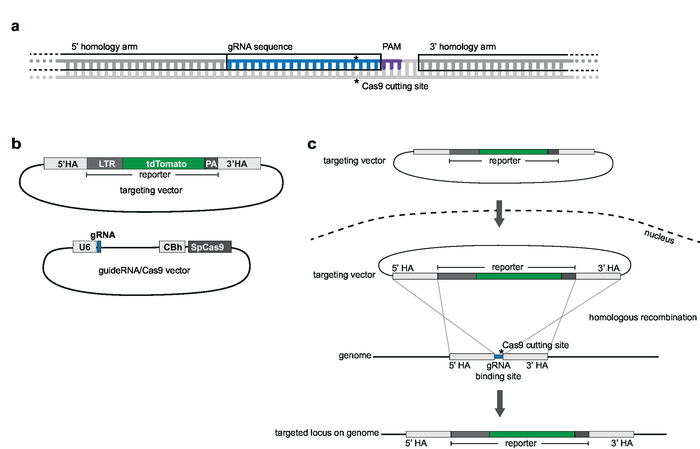
Figure 2: Targeting strategy and vector design. (a) gRNA and choice of homology arms. 20 nt gRNA is homologous to the chosen genomic target site and situated adjacent to a PAM. Homology arms are complementary to 1,000 bp up- and downstream of the gRNA and should not include the gRNA sequence. (b) Schematics of targeting vector and gRNA/Cas9 vector. The targeting vector consists of the chosen reporter sequence that is 5' and 3' flanked by the homology arms. The gRNA/Cas9 vector is based on the pX330-U6-Chimeric_BB-cBh-hSpCas9 backbone. (c) Schematic of targeting by homologous recombination. Target vector and guideRNA/Cas9 vector are transfected into Jurkat cells. Cas9 mediates a double strand break at genomic target site (indicated by *) and facilitates homologous recombination and integration of reporter sequence into the genomic target locus. Please click here to view a larger version of this figure.
2. CRISPR-Cas9-Based Targeting of Jurkat Cells
- Transfection of Jurkat cells
- 24 h prior to transfection, plate 1.25 x 106 Jurkat T cells in 2.5 mL of RPMI 1640 supplemented with 10% (v/v) fetal calf serum (FCS) and 4 mM L-glutamine [referred to as “RPMI w.o. antibiotics (AB)”] per well of a 6-well cell culture plate. For a single targeting experiment, prepare one complete 6-well plate (i.e., 6 wells each with 2.5 mL of cell suspension).
- On the following day, co-transfect the cells with circular tv and pX330-U6-Chimeric_BB-cBh-hSpCas9/gRNA using a transfection reagent specific for Jurkat cells.
- Add 2 µg of circular tv and 2 µg of pX330-U6-Chimeric_BB-cBh-hSpCas9/gRNA per well to 250 µL of commercial RPMI medium with reduced serum concentration optimized for transfection (RPMI with 50% reduction in serum) in a reaction tube and mix well.
- Add 12 µL of transfection reagent slowly to the DNA/medium without touching the wall of the tube and swirl. Let the mixture incubate for 15 min and add dropwise to one well of cells. Incubate the cells at 37 °C and 5% CO2.
NOTE: The preparation of transfection reaction can be scaled up. No medium change is required after transfection.
- Enrichment of transfected cells by fluorescence-activated cell sorting (FACS)
- 72 h post-transfection, pool the transfected cells, count them, and prepare for enrichment by FACS. Collect the cells in a 50 mL conical tube, centrifuge at 300 x g and room temperature (RT) for 4 min, wash the cells once in PBS, centrifuge again, suspend the pellet in an appropriate amount of FACS buffer (PBS supplemented with 1% FCS + 1 mM EDTA) at 1 x 107 cells/mL, and finally transfer into a FACS tube.
- Subject the cells to FACS and sort those that express the fluorescent reporter of the tv (e.g., tdTomato in this protocol). Collect the cells in RPMI 1640 supplemented with 10% FCS, 4 mM L-glutamine, and 50 U/mL penicillin and streptomycin (referred to as “RPMI w/ AB”).
- After FACS sorting, wash the cells once by adding 20 mL of RPMI w/ AB to sorted cells and centrifuge at 300 x g for 4 min at RT. Resuspend the cell pellet in an appropriate amount of RPMI w/ AB and plate the cells in one well of a cell culture plate with the appropriate volume according to cell number post-FACS.
NOTE: Culture sort the cells in a small volume (e.g., 24-well), as considerable levels of cell death have been observed in the first week post-targeting (up to 80–90%). - Expand the mixed targeted cell population up to a density of 1 x 106 cells/mL in a 75 cm² cell culture flask. This will take around 10–14 days post-FACS sorting.
- Confirmation of targeting events by flow cytometry in the mixed targeted cell population
- After 10–14 days of expansion (when cells have reached a density of 1 x 106 cells/mL in a 75 cm2 cell culture flask), plate two wells with 1 x 106 cells of the mixed targeted cell population in 1 mL of RPMI w/ AB in a 12-well cell culture plate.
- Induce the HIV LTR of the reporter (generation of tv is described in step 1.2.2.1.) in one of the wells by adding 50 ng/mL phorbol 12-myristate 13-acetate (PMA) and 1 µM Ionomycin (referred to as PMA-Iono). Use cells in the second well as the non-induced control. Culture both the induced and the non-induced cells for 24 h.
- Take 0.5 mL of cell suspension of non-induced and induced cells (each), wash them once in PBS, and suspend each in 200 µL of FACS buffer.
- Analyze 100,000 cells by flow cytometry. Gate the viable single-cells based on size in forward and sideward scatter, and analyze fluorescent reporter gene expression.
NOTE: At this step (10–14 days post-FACS sorting), transient expression of fluorescent reporter by transfection should no longer be detectable. Fluorescent expression at this time point indicates genomic integration of reporter sequence.
- Detection of targeting events by PCR on genomic DNA of the mixed targeted cell population
NOTE: To detect targeting events via PCR, design two primer pairs specific for the 5’ integration (int.) junction and 3’ int. junction. For the 5’ int. junction PCR, the forward primer should bind upstream of the 5’ HA and the reverse primer in the LTR of the reporter (primers P1 and P2 in Figure 3a). The primer pair for the 3’ int. junction PCR should span from the PA of the reporter to 100–200 bp downstream of the 3’ HA (primers P3 and P4 in Figure 3a). Primers P1 and P4 will also serve for amplification of the non-targeted allele in the mixed targeted population. For a schematic, see Figure 3a.- Prepare gDNA from 2 mL of cell suspension of the mixed targeted cell population from step 2.2.4. Use a gDNA extraction kit according to the manufacturer’s protocol. Then prepare the gDNA of non-targeted cells as a control.
- Perform int. junction PCRs (primers P1/P2 and P3/P4 for 5’ and 3’ int. junction, respectively) and non-targeted allele PCR (primers P1/P4) using a high-fidelity DNA polymerase (see Tables 3 and 4 for PCR ingredients and cycling conditions). Analyze 5 µL of PCR products on a 1.5% agarose/TAE gel.
NOTE: If the mixed targeted population contains cells that have undergone genome engineering, a specific PCR product should be observed that is not detectable in gDNA of non-targeted cells (negative control). For the non-targeted allele PCR, one should observe a product of the same size for both the targeted and non-targeted cells (positive control for genomic P1 and P4 primers). If no bands are observed, consider altering PCR cycling conditions by increasing the number of cycles or altering the PCR buffer (for example through addition of DMSO or increased amounts of Mg2+), or by changing the polymerase.
3. Generation of Clonal Lines and Screening for Correct Targeting
NOTE: After confirmation of the targeting events in the mixed targeted cell population by flow cytometry and PCR (sections 2.2–2.4), generate single-cell clones (duration: 28 to 35 days) and screen for correct integration of the reporter sequence.
- Generation of single-cell clones through dilution plating
- Prepare Jurkat-conditioned medium in advance: take off RPMI w/ AB medium from healthy, untreated Jurkat T cells grown to 1 x 106 cells/mL, centrifuge for 5 min at 300 x g, and filter the supernatant using a 0.22 µm syringe filter unit.
NOTE: Keep the conditioned medium at 4 °C for short-term storage or at -20 °C for storage longer than 1 week. Prepare 20 to 30 mL of conditioned medium prior to dilution plating. - Count the targeted cells from step 2.2.4 after 10–14 days of expansion and dilute them in RPMI w/ AB to a concentration of 1 x 105 cells/mL. Take 100 µL of 1 x 105 cells/mL solution and dilute with 9.9 mL of medium to achieve a concentration of 1,000 cells/mL. Take 1 mL of 1,000 cells/mL solution and dilute with 9 mL of medium to achieve a concentration of 100 cells/mL.
- Plate 96-well plates containing 1 cell per well and 2 cells per well. For 1 cell per well, take 1 mL of 100 cells/mL solution and mix gently with 5 mL of conditioned medium and 4 mL of fresh medium in a sterile reagent reservoir.
- For 2 cells per well, take 2 mL of 100 cells/mL solution and mix gently with 5 mL of conditioned medium and 3 mL of fresh medium. Plate 96-well round-bottom plates with 100 µL of the respective cell dilution per well using a multichannel pipet.
NOTE: 5 to 10 96-well plates per targeting construct are sufficient to obtain clones for screening. - Stack the 96-well plates, cover each stack with a 6-well plate containing 3 mL of PBS in each well, and incubate the plates at 37 °C in a humidified cell culture incubator with 5% CO2 for 3 weeks.
NOTE: Do not change the cell culture medium during this time. Do not open the incubator more than once or twice a week. The best results are observed in incubators with open water reservoir. - After 3 weeks of incubation, visually confirm the presence of grown colonies using light microscopy (4X magnification) and mark the wells with grown colonies so they are visible as points on the bottom of the wells.
- Prepare one 96-well round-bottom plate with 100 µL of RPMI w/ AB per well. Gently resuspend cells of a marked well by pipetting. Transfer 100 µL of cell suspension into one well of the new 96-well plate already containing 100 µL of RPMI w/ AB, then mix gently by pipetting. Transfer 100 µL of this cell suspension into a second empty 96-well round-bottom plate to duplicate the plate.
- Continue with all marked wells with grown colonies. Fill all the blank wells with 200 µL of RPMI w/ AB medium. Incubate the plates at 37° C and 5% CO2.
NOTE: One of these plates will serve for expansion of single-cell clones (“stock plate”) and the other as a “duplicate plate” for screening.
- Prepare Jurkat-conditioned medium in advance: take off RPMI w/ AB medium from healthy, untreated Jurkat T cells grown to 1 x 106 cells/mL, centrifuge for 5 min at 300 x g, and filter the supernatant using a 0.22 µm syringe filter unit.
- Screening of single-cell clones by flow cytometry and PCR
NOTE: While the single-cell clones are expanding, use the duplicate plate from step 3.1.8 to screen single-cell clones for presence of reporter sequence by PCR (steps 3.2.4–3.2.12) and expression of fluorescent reporter by flow cytometry (steps 3.2.2–3.2.3) (Figure 3c).- Let the duplicate plate incubate for 24 to 48 h and duplicate the plate again. To do this, add 100 µL of RPMI w/ AB to every well, mix gently by pipetting, and transfer 100 µL to a new 96-well round-bottom plate using a multichannel pipet. Use one plate for flow cytometry screening and the other for PCR-based screening.
- For flow cytometry screening, stimulate the cells with PMA-Iono. Prepare a mastermix of 0.1 µL of Ionomocin (1 mM stock), 0.1 µL of PMA (50 µg/µL stock), and 4.8 µL of RPMI w/ AB per number of wells, then add 5 µL of mastermix per well.
NOTE: Induction is necessary to successfully identify clones, where the LTR might be transcriptionally silent and therefore the fluorescent reporter is not expressed. - Let cells incubate for 24 h and prepare cells for flow cytometry as described in step 2.3.3. Gate any viable single-cells based on size in forward and sideward scatter and analyze fluorescent reporter gene expression by flow cytometry (for example results, see Figure 3c). If tv backbone contains a second fluorescent reporter with promoter (e.g., GFP), screen any clones for backbone reporter expression also (see step 1.2.2 and the following note for explanation).
NOTE: Backbone reporter expression indicates unwanted integration of backbone sequences. - Once the clones in the second duplicate plate have grown sufficiently (usually 24 to 48 h after duplication of the 96-well plate), prepare cell lysates containing gDNA for PCR screening. Centrifuge the plate for 10 min at 300 x g at RT. Carefully take off the supernatant without disturbing the cell pellet.
NOTE: All steps for the preparation of lysates and PCR reactions can be performed with multichannel pipettes. - Wash cells with 100 µL of PBS by gentle pipetting and centrifuging the plate for 5 min at 300 x g at RT. Take off the PBS and add 200 µL of lysis buffer [200 mM NaCl, 100 mM Tris-HCl pH 8-8.5, 5 mM EDTA, 0.1% SDS; then add 250–1,000 µg/mL of proteinase K (lyophilized powder, weigh in freshly)] per well. Mix gently by pipetting, and transfer the suspension to a new PCR plate.
- Seal the plate with paraffin film and incubate for 1 h at 55 °C in a thermocycler. Centrifuge at maximum speed for 10 min (3,000 x g) to spin down cell debris, and transfer the supernatant to a new PCR plate.
NOTE: Cell lysates in plates can be stored at this stage at 4 °C until further use. - Prepare a 96-well PCR plate with 110 µL of dH2O and add 10 µL of cell lysate (1:12 dilution). Cell lysates might be viscous and difficult to pipet. Use at least 20-µL pipet tips.
- Inactivate proteinase K by incubation for 10 min at 99 °C in a thermocycler. Subsequently use the inactivated and diluted cell lysates for PCR screening.
- Design primers for screening PCR (P5 and P6) based on the chosen reporter sequence to amplify 500–800 bp of the reporter sequence. For positive control PCR, use primers P7 and P8 that amplify 630 bp of a wild-type, non-targeted genomic locus (NUP188 gene) (Figure 3c and Table 5). Design a third primer pair that amplifies 500–600 bp of the tv backbone as a control for unspecific integration of tv backbone sequences (backbone PCR).
- For screening, control, and backbone PCR, use a commercial PCR mastermix (see Tables 6 and 7 for PCR ingredients and cycling conditions). Use 2 µL of the diluted and inactivated lysate prepared in step 3.2.8 as a template and run PCR for 38 to 40 cycles of PCR amplification in 96-well format.
- Analyze 5 µL of PCR products on a 1.5% agarose/TAE gel.
NOTE: For control PCR, a specific band of 630 bp should be observed for every sample, confirming that the quality of cell lysates is adequate for PCR. A specific band in screening PCR (500–800 bp depending on primer design) indicates integration of the reporter sequence. A specific band for backbone PCR (500–600 bp, depending on primer design) indicates unwanted integration of tv backbone sequences (for example results, see Figure 3c). - Combine the results of flow cytometry (step 3.2.3) and PCR-based screening (step 3.2.12). Select clones which show correct sizes of PCR products in screening PCR and positive control PCR and expression of fluorescent reporter after induction with PMA-Iono in flow cytometry. Exclude clones that show any PCR product in backbone PCR or expression of tv backbone-encoded fluorescent protein, indicating unspecific integration of tv backbone sequence.
- Gradually expand selected clones from the 96-well stock plate to bigger well formats (48/24/12/6-well) until achieving a T75 cell culture flask format by adding fresh medium every 2 to 3 days. Maintain a cell density between 1 x 105 and 1 x 106 cells/mL.
- Make sure to prepare cell stocks of clones during expansion: count the cells, centrifuge at 300 x g for 5 min at RT, discard the supernatant, and suspend the cells gently in FCS + 10% DMSO at 5 x 106 cells/mL. Aliquot in cryogenic vials and use a cryo-freezing container to freeze the cells to 80 °C at 1 °C/min. For long term storage, transfer them to liquid N2.
NOTE: It is advisable to retain a T75 cell culture flask (i.e., around 1 x 107 cells) during the expansion in preparation of gDNA for verification of targeting by Southern blotting (see section 3.4).
- Verification of integration sites by PCR/sequencing in selected clones
NOTE: 5’ and 3’ int. junctions of the selected clones are PCR amplified and submitted to Sanger sequencing to verify correct targeting at the DNA sequence level.- Prepare gDNA of the selected clones and Jurkat wild-type cells using a commercial gDNA extraction kit.
- Use primer pairs binding the 5’ end of the reporter and upstream of 5’ HA for 5’ int. junction (primers P1 and P2) and the 3’ end of the reporter and downstream of 3’ HA for 3’ int. junction (primers P3 and P4) as described in step 2.4. Use primers P1 and P4 to amplify the targeted integration site on the allele without reporter integrant (Figure 4a).
- Prepare PCR reactions with 100–200 ng of gDNA as a template and perform PCR using a polymerase with proofreading activity (see Tables 1 and 2 for PCR ingredients and cycling conditions).
NOTE: If no bands are observed, consider altering the PCR cycling conditions by increasing the number of cycles or altering the PCR buffer (for example, by adding DMSO or increased amounts of Mg2+), or by changing the polymerase. - Analyze 5 µL of PCR products on a 1.5% agarose/TAE gel. If correct band sizes are observed, purify the remaining PCR product using a commercial kit and subject it to Sanger sequencing. Verify sequences of 5’ int. junction, 3’ int. junction, and targeted site of the allele without reporter integrant by aligning them with expected sequences.
NOTE: The homologous allele where the reporter has not integrated will likely show Cas9-mediated changes at the target site, such as nucleotide insertions or deletions (Figure 4a). - For clones that show correct int. junction sequences after alignment, perform a PCR amplifying the whole targeted reporter and subject it to Sanger sequencing to verify the correct sequence of the integrant.
- Southern blot analysis for verification of targeting in selected clones
NOTE: Southern blot analysis of selected clones is required to verify correct targeting and exclude Cas9-mediated recombination events that may have occurred at the targeted integration site.- Develop a strategy for appropriate gDNA digestion and probe the design prior to starting the experiment.
- Select a restriction enzyme for gDNA restriction that generates appropriate fragments of 2 to 10 kb length at the targeted site. Certain restriction enzymes, such as Asp718, BamHI, BglI, BglII, EcoRV, HindIII, NcoI, PstI, PvuII, ScaI, StuI, and SstI have been successfully used for digesting high molecular weight gDNA.
- Design two different Southern probes: a reporter-specific probe and genomic probe. The reporter-specific probe hybridizes to a sequence within the reporter (i.e., tdTomato-specific probe). The genomic probe hybridizes to a genomic region close to (but not overlapping) one HA.
- Choose the genomic probe so that the fragment generated by gDNA digestion that will be detected by genomic probe binding differs in length (more than 2 kb) from the targeted and non-targeted alleles (Figure 4b). A probe length of 400 to 1,000 bp is recommended.
- Design PCR primers to amplify the two required probes. Amplify the reporter-specific probe from tv template using a high-fidelity DNA polymerase (see Tables 3 and 4 for PCR ingredients and cycling conditions).
- Amplify the genomic probe from wild-type Jurkat gDNA prepared with a commercial gDNA extraction kit using a DNA polymerase with proofreading activity (see Tables 1 and 2 for PCR ingredients and cycling conditions). Purify the PCR products on an agarose/TAE gel and extract the fragments using a commercial gel extraction kit according to the manufacturer’s instructions.
- Extract high molecular weight gDNA from 1 x 107 cells of the wild-type Jurkat cells and selected clones from step 3.2.14.
- Pellet the cells by centrifugation at 300 x g for 5 min at RT, wash it once with PBS, and suspend the pellet in 4 mL of lysis buffer [200 mM NaCL, 100 mM Tris-HCl pH 8, 5 mM EDTA, 0.1% SDS; then add 250–1,000 µg/mL proteinase K (lyophilized powder, weigh in freshly)]. Incubate o/n at 55 °C, shaking at 350 rpm in a tabletop thermomixer.
- Add 4 mL of isopropanol and mix by inversion 10 to 20 times. The gDNA should become visible as white precipitate. Spool the precipitated gDNA onto the fine tip of a glass pipette, wash by emerging in 750 µL of 70% EtOH, and let dry at RT (5 to 10 min).
- Shed the precipitate into a 1.5 mL reaction tube containing 500 µL of 1x TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA) and leave to dissolve o/n at 4 °C, shaking at 350 rpm. Any pipetting of gDNA from this stage should be done with wide-bore tips to avoid shearing.
NOTE: Preparation of high molecular weight gDNA is essential for Southern blot analysis, and commercially available gDNA preparation kits are not suitable.
- Digest (two times) 15 µg of gDNA of the selected clones and wild-type Jurkat cells with selected restriction enzyme (see step 3.4.1.1) in a 60 µL reaction with 6 µL of enzyme (20 Units/µL): first, add DNA, digestion buffer, and ddH2O, incubate o/n at 37 °C, then add enzyme and incubate o/n at enzyme-specific digestion temperature. 15 ug of digested gDNA is required per Southern probe.
- Use 7 µL of the 60 µL restriction digest for analytical gel electrophoresis on a 1% agarose/TAE gel. A smear indicates complete digestion and good DNA quality for Southern blot analysis.
- Precipitate the remaining restriction digest by adding 1:10 3 M sodium acetate and 2 volumes 100% EtOH, then incubate for 1 h at -80 °C and centrifuge for 30 min at 15,600 x g at 4 °C.
- Discard the supernatant and wash the pellet with 70% EtOH. Centrifuge for 15 min at 15,600 x g at 4 °C, discard the supernatant, let the pellet dry briefly at RT, and dissolve in 20 µL of ddH2O.
- Run 1% agarose/TAE blotting gel, loading 20 uL of digested gDNA per lane. Run the gel for 2 h at 60 V, 400 mA.
NOTE: The percentage of agarose gel and running time/voltage may be adjusted according to expected fragment size for Southern blot detection calculated in step 3.4.1.1. The following steps of Southern blot analysis are described in detail in a supplementary protocol (steps 1 to 18). These steps comprise of: washing of the blotting gel, blotting onto a nylon membrane, radioactive probe generation, probe hybridization, and development of autoradiograph film. Compare the obtained banding pattern after autoradiograph development in step 18 (supplementary protocol) with the expected pattern according to Southern strategy (for example results, see Figure 4b).
- Develop a strategy for appropriate gDNA digestion and probe the design prior to starting the experiment.
- Analysis of off-target events
NOTE: Since CRISPR-Cas9-mediated genome engineering can generate off-target effects, PCR-amplify the ten highest-ranked in silico-predicted off-target sites in selected clones and subject them to Sanger sequencing.- Use CCTop16 (http://crispr.cos.uni-heidelberg.de) to generate a list of the ten highest-ranked in silico predicted off-target sequences.
- Input the gRNA sequence including PAM as used for targeting as the query sequence. Select “NGG” as PAM and “Human genome” as the reference for off-target prediction.
- Set maximal total mismatches to “4” and target site length to the length of the gRNA without PAM. The output file will provide a ranked list of genomic off-target sites for respective gRNA.
- In silico extract the genomic sequence 500 bp upstream and downstream of each of the ten highest-ranked off-target hits using UCSC Genome Browser (http://genome.edu.ucsc.edu) and the position of the off-target hit from CCTop results list.
- For each off the target sites to be analysed, design a PCR primer pair that amplifies a fragment of 600 to 700 bp in length including the predicted off-target site.
- Extract gDNA from the selected clones and Jurkat wild-type cells using a commercial gDNA extraction kit. For every off-target site, perform a PCR using a DNA polymerase with proofreading activity (see Tables 1 and 2 for PCR ingredients and cycling conditions) on wild-type and the respective clone-derived gDNA.
- Analyze 5 µL of PCR products on a 1.5% agarose/TAE gel. If correct band sizes are observed, purify the remaining PCR product using a commercial PCR purification kit and subject it to Sanger sequencing. Compare sequences of the off-target sites in Jurkat cells and the targeted clones.
- Use CCTop16 (http://crispr.cos.uni-heidelberg.de) to generate a list of the ten highest-ranked in silico predicted off-target sequences.
Results
In this representative experiment we have chosen to target a minimal HIV-1-derived reporter consisting of a LTR, tdTomato-coding sequence, and polyA-signal sequence to two loci in intron 5 of the BACH2 gene17. The loci for targeting were chosen according to proximity to published recurrent integration sites found in different studies on primary T cells from HIV-infected patients2,4,
Discussion
Here, we describe a protocol to generate HIV-1-derived Jurkat reporter models with chosen proviral integration sites applying CRISPR-Cas9-based genome engineering.
Several points of the protocol require careful attention during the planning stage. First, the locus to be targeted should be chosen carefully, as some loci might be easier to target than others (e.g., depending on the chromatin status of the region and the target sequence itself). Repetitive sequences are hard to clone int...
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Britta Weseloh and Bettina Abel for technical assistance. We also thank Arne Düsedau and Jana Hennesen (flow cytometry technology platform, Heinrich Pette Institut) for technical support.
Materials
| Name | Company | Catalog Number | Comments |
| pX330-U6-Chimeric_BB-cBh-hSpCas9 | Addgene | 42230 | vector for expression of SpCas9 and gRNA |
| pMK | GeneArt | mammalian expression vector for cloning | |
| cDNA3.1 | Invitrogen | V79020 | mammalian expression vector for cloning |
| BbsI | New England Biolabs | R0539S | restriction enzyme |
| NEBuilder Hifi DNA Assembly Cloning Kit | New England Biolabs | E5520S | Assembly cloning kit used for target vector generation |
| TaqPlus Precision PCR System | Agilent Technologies | 600210 | DNA polymerase with proofreading activity used for amplification of homology arms (step 1.2.2.2), verification of integration site and reporter sequence (step 3.3.3 and 3.3.5), generation of genomic probe for Southern blot (step 3.4.1.5) and analysis of off-target events (step 3.5.4) |
| 96-well tissue culture plate (round-bottom) | TPP | 92097 | tissue culture plates for dilution plating |
| Phusion High-Fidelity DNA polymerase | New England Biolabs | M0530 L | DNA polymerase used for detection of targeting events (step 2.4.2) and generation ofreporter-specific probe for Southern blot (step 3.4.1.4) |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D9170 | dimethyl sulfoxide as PCR additive |
| Magnesium Chloride (MgCl2) Solution | New England Biolabs | B9021S | MgCl2 solution as PCR additive |
| Deoxynucleotide (dNTP) Solution Mix | New England Biolabs | N0447S | dNTP mixture with 10 mM of each nt for PCR reactions |
| 5PRIME HotMasterMix | 5PRIME | 2200400 | ready-to-use PCR mix used for screening PCR (step 3.2.11) |
| QIAamp DNA blood mini kit | Qiagen | 51106 | DNA isolation and purification kit |
| QIAquick PCR Purification Kit | Qiagen | 28106 | PCR Purification Kit |
| RPMI 1640 without glutamine | Lonza | BE12-167F | cell culture medium |
| Fetal Bovine Serum South Africa Charge | PAN Biotech | P123002 | cell culture medium supplement |
| L-glutamine | Biochrom | K 0282 | cell culture medium supplement |
| Penicillin/Streptomycin 10.000 U/mL/ 10.000 µg/mL | Biochrom | A 2212 | cell culture medium supplement |
| Gibco Opti-MEM Reduced Serum Media | Thermo Fisher Scientific | 31985062 | cell culture medium with reduced serum concentration optimized for transfection |
| TransIT-Jurkat | Mirus Bio | MIR2125 | transfection reagent |
| phorbol 12-myristate 13-acetate | Sigma-Aldrich | P8139-1MG | cell culture reagent |
| Ionomycin | Sigma-Aldrich | I0634-1MG | cell culture reagent |
| Syringe-driven filter unit, PES membrane, 0,22 µm | Millex | SLGP033RB | filter unit for sterile filtration |
| Heracell 150i incubator | Thermo Fisher Scientific | 51026280 | tissue culture incubator |
| Amershan Hybond-N+ | GE Healthcare | RPN1520B | positively charged nylon membrane for DNA and RNA blotting |
| Stratalinker 1800 | Stratagene | 400072 | UV crosslinker |
| High Prime | Roche | 11585592001 | kit for labeling of DNA with radioactive dCTP using random oligonucleotides as primers |
| illustra ProbeQuant G-50 Micro Columns | GE Healthcare | 28-9034-08 | chromatography spin-columns for purification of labeled DNA |
References
- Hughes, S. H., Coffin, J. M. What Integration Sites Tell Us about HIV Persistence. Cell Host and Microbe. 19 (5), 588-598 (2016).
- Marini, B., Kertesz-Farkas, A., et al. Nuclear architecture dictates HIV-1 integration site selection. Nature. 521 (7551), 227-231 (2015).
- Cesana, D., Santoni de Sio, F. R., et al. HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells. Nature Communications. 8 (1), 498 (2017).
- Cohn, L. B., Silva, I. T., et al. HIV-1 Integration Landscape during Latent and Active Infection. Cell. 160 (3), 420-432 (2015).
- Han, Y., Lassen, K., et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. Journal of Virology. 78 (12), 6122-6133 (2004).
- Ikeda, T., Shibata, J., Yoshimura, K., Koito, A., Matsushita, S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. The Journal of Infectious Diseases. 195 (5), 716-725 (2007).
- Wagner, T. A., Mclaughlin, S., et al. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 345 (6196), 570-573 (2014).
- Maldarelli, F., Wu, X., et al. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 345 (6193), 179-183 (2014).
- Jordan, A., Bisgrove, D., Verdin, E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. The EMBO Journal. 22 (8), 1868-1877 (2003).
- Mack, K. D., Jin, X., et al. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. Journal of Acquired Immune Deficiency Syndromes. 33 (3), 308-320 (2003).
- Byrne, S. M., Ortiz, L., Mali, P., Aach, J., Church, G. M. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Research. 43 (3), 1-12 (2014).
- CRISPR Genome Engineering Toolbox: Target Sequence Cloning Protocol. Addgene website Available from: https://www.addgene.org/static/cms/filer_public/e6/5a/e65a9ef8-c8ac-4f88-98da-3b7d7960394c/zhang-lab-general-cloning-protocol.pdf (2013)
- Gibson Assembly Protocol. Addgene website Available from: https://www.addgene.org/protocols/gibson-assembly/ (2009)
- Addgene Plasmid Cloning by PCR. Addgene website Available from: https://www.addgene.org/protocols/pcr-cloning/ (2014)
- Addgene Plasmid Cloning by Restriction Enzyme Digest (aka Subcloning). Addgene website Available from: https://www.addgene.org/protocols/subcloning/ (2013)
- Stemmer, M., Thumberger, T., Del Sol Keyer, M., Wittbrodt, J., Mateo, J. L. CCTop: An intuitive, flexible and reliable CRISPR-Cas9 target prediction tool. Public Library of Science (PLoS) ONE. 10 (4), (2015).
- Lange, U. C., Bialek, J. K., Walther, T., Hauber, J. Pinpointing recurrent proviral integration sites in new models for latent HIV-1 infection. Virus Research. 249, (2018).
- Bialek, J. K., Dunay, G. A., et al. Targeted HIV-1 Latency Reversal Using CRISPR-Cas9-Derived Transcriptional Activator Systems. PloS ONE. 11 (6), e0158294 (2016).
- Lee, C. M., Davis, T. H., Bao, G. Examination of CRISPR-Cas9 design tools and the effect of target site accessibility on Cas9 activity. Experimental Physiology. 103 (4), 456-460 (2018).
- Jensen, K. T., Fløe, L., et al. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Letters. 591 (13), 1892-1901 (2017).
- Simonetti, F. R., Sobolewski, M. D., et al. Clonally expanded CD4 + T cells can produce infectious HIV-1 in vivo. Proceedings of the National Academy of Sciences. 113 (7), 1883-1888 (2016).
- Chen, H. C., Martinez, J. P., Zorita, E., Meyerhans, A., Filion, G. J. Position effects influence HIV latency reversal. Nature Structural and Molecular Biology. 24 (1), 47-54 (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved